3P8B
 
 | |
6RNZ
 
 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
8TQS
 
 | |
4A0A
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
3PMN
 
 | | ternary crystal structure of polymerase lambda variant with a GT mispair at the primer terminus with Mn2+ in the active site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 5'-D(*CP*AP*GP*TP*AP*G)-3', 5'-D(*CP*GP*GP*CP*CP*TP*TP*AP*CP*TP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7AT8
 
 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | Deposit date: | 2020-10-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
7KUI
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
3PML
 
 | | crystal structure of a polymerase lambda variant with a dGTP analog opposite a templating T | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 5'-D(*CP*AP*GP*TP*AP*C)-3', 5'-D(*CP*GP*GP*CP*TP*GP*TP*AP*CP*TP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8XPB
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Huang, H.T, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
1Q1V
 
 | |
8C45
 
 | |
6Y5E
 
 | | Structure of human cGAS (K394E) bound to the nucleosome (focused refinement of cGAS-NCP subcomplex) | | Descriptor: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-C, ... | | Authors: | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
5T2O
 
 | |
5T2H
 
 | |
5T2N
 
 | |
6LKE
 
 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
8HHL
 
 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA full R-loop complex | | Descriptor: | Cas12m2, MAGNESIUM ION, NTS (36-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1FLO
 
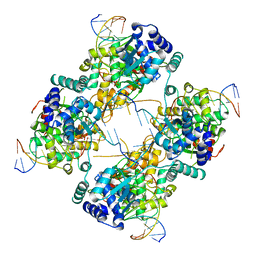 | | FLP Recombinase-Holliday Junction Complex I | | Descriptor: | FLP RECOMBINASE, PHOSPHONIC ACID, SYMMETRIZED FRT DNA SITES | | Authors: | Chen, Y, Narendra, U, Iype, L.E, Cox, M.M, Rice, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping.
Mol.Cell, 6, 2000
|
|
1K3X
 
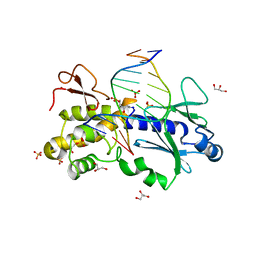 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
6XJD
 
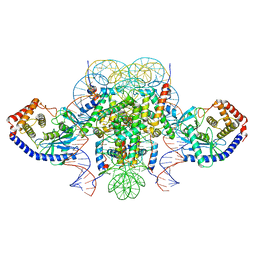 | | Two mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Xu, P, Li, P, Zhao, B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
5A72
 
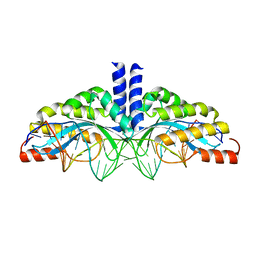 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
7BZG
 
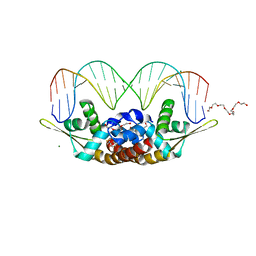 | | Structure of Bacillus subtilis HxlR, wild type in complex with formaldehyde and DNA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*GP*TP*AP*TP*CP*CP*TP*CP*GP*AP*GP*GP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
6X5A
 
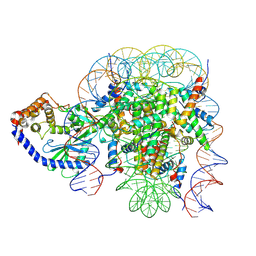 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
8IIR
 
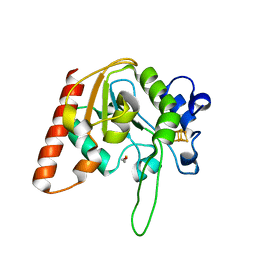 | | MsmUdgX H109S/Q53A double mutant | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
1C3J
 
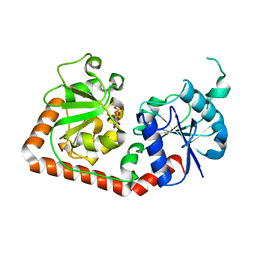 | | T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, A, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism.
J.Mol.Biol., 292, 1999
|
|
