2VS4
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2VS5
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
5HDP
 
 | | Hydrolase StnA mutant - S185A | | Descriptor: | Hydrolase, methyl 5-amino-6-(7-amino-6-methoxy-5,8-dioxo-5,8-dihydroquinolin-2-yl)-4-(2-hydroxy-3-methoxyphenyl)-3-methylpyridine-2-carboxylate | | Authors: | Qian, T. | | Deposit date: | 2016-01-05 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of StnA for the Biosynthesis of Antitumor Drug Streptonigrin Reveals a Unique Substrate Binding Mode
Sci Rep, 7, 2017
|
|
5HDF
 
 | | Hydrolase SeMet-StnA | | Descriptor: | Hydrolase | | Authors: | Qian, T. | | Deposit date: | 2016-01-05 | | Release date: | 2017-01-11 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of StnA for the Biosynthesis of Antitumor Drug Streptonigrin Reveals a Unique Substrate Binding Mode
Sci Rep, 7, 2017
|
|
1NBO
 
 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
5JHK
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-6 ligand. | | Descriptor: | N-(benzenecarbonyl)glycyl-L-arginine, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5J1X
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-5 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(tert-butoxycarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5KIT
 
 | |
5JGI
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with M45 compound | | Descriptor: | N-ALPHA-L-ACETYL-ARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5JGQ
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-7 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(benzenecarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5IYY
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-4 ligand. | | Descriptor: | Neuropilin-1, N~2~-[(benzyloxy)carbonyl]-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
7P2O
 
 | |
6LKN
 
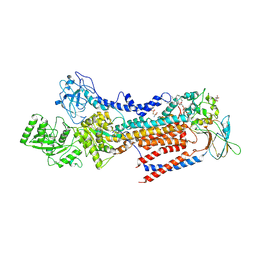 | | Crystal structure of ATP11C-CDC50A in PtdSer-bound E2P state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Hasegawa, K. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of a human plasma membrane phospholipid flippase.
J.Biol.Chem., 295, 2020
|
|
6M9C
 
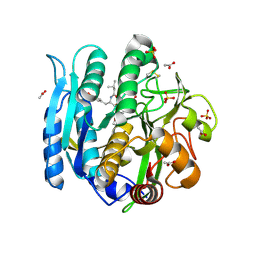 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Pseudotyrostatin | | Descriptor: | ACETIC ACID, CALCIUM ION, Pseudotyrostatin, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
5CF3
 
 | | Crystal structures of Bbp from Staphylococcus aureus | | Descriptor: | Bone sialoprotein-binding protein, CALCIUM ION | | Authors: | Yu, Y, Zhang, X.Y, Gu, J.K. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen alpha
Protein Cell, 6, 2015
|
|
5CFA
 
 | |
4AJB
 
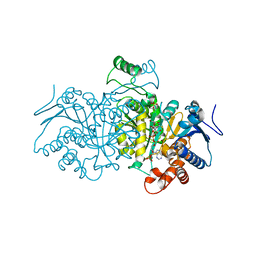 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with Isocitrate, magnesium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJS
 
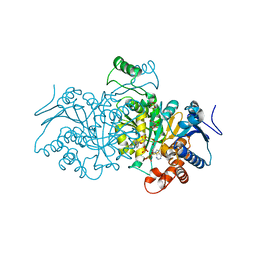 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate, magnesium(II), Adenosine 2',5'-biphosphate and ribosylnicotinamide-5'-phosphate | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJR
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, magnesium(II) and NADPH - The product complex | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJC
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, calcium(II) and adenine nucleotide phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-2'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJ3
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and NADP - The pseudo-Michaelis complex | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP ISOCITRATE DEHYDROGENASE, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJA
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | Descriptor: | Outer membrane protein, octyl beta-D-glucopyranoside | | Authors: | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
4YLS
 
 | | Tubulin Glutamylase | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tubulin polyglutamylase TTLL7 | | Authors: | Garnham, C.P, Vemu, A, Wilson-Kubalek, E.M, Yu, I, Szyk, A, Lander, G.C, Milligan, R.A, Roll-Mecak, A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases.
Cell, 161, 2015
|
|
4YLR
 
 | | Tubulin Glutamylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Tubulin polyglutamylase TTLL7 | | Authors: | Garnham, C.P, Vemu, A, Wilson-Kubalek, E.M, Yu, I, Szyk, A, Lander, G.C, Milligan, R.A, Roll-Mecak, A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases.
Cell, 161, 2015
|
|
