2VSF
 
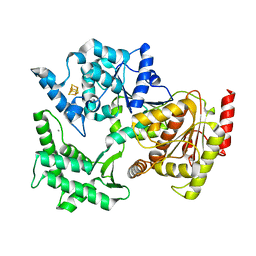 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
6WVC
 
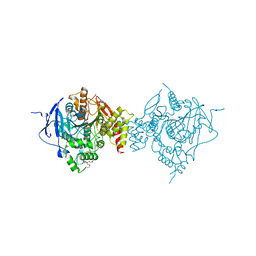 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GD | | Descriptor: | (1S)-2,2-dimethylcyclopentyl (R)-methylphosphinate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WVP
 
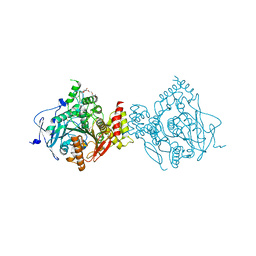 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GF | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUY
 
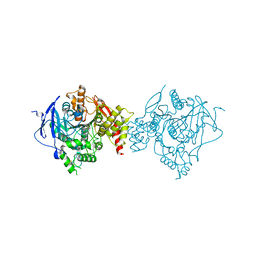 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GA and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUV
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WV1
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GB and HI-6 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUZ
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GB | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
2WKR
 
 | | Structure of a photoactivatable Rac1 containing the Lov2 C450M Mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
2WKY
 
 | | Crystal structure of the ligand-binding core of GluR5 in complex with the agonist 4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Glutamate Receptor Glur5 Agonist (S)-2-Amino-3-(3-Hydroxy-7,8-Dihydro-6H-Cyclohepta[D]Isoxazol-4-Yl)Propionic Acid and the 8-Methyl Analogue: Synthesis, Molecular Pharmacology, and Biostructural Characterization
J.Med.Chem., 52, 2009
|
|
6WVO
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GD and HI-6 | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
2WKP
 
 | | Structure of a photoactivatable Rac1 containing Lov2 Wildtype | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
8JZ7
 
 | | Cryo-EM structure of MK-6892-bound HCAR2 in complex with Gi protein | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-07-04 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric ligand selectivity and allosteric probe dependence at Hydroxycarboxylic acid receptor HCAR2.
Signal Transduct Target Ther, 8, 2023
|
|
8K5C
 
 | |
8K5B
 
 | |
8K5D
 
 | |
8KG8
 
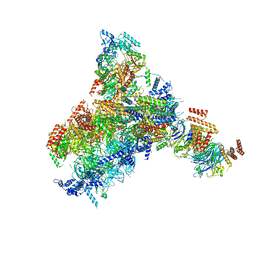 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG6
 
 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG9
 
 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
2WJY
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 Orthorhombic form | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, SULFATE ION, ZINC ION | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
2E5D
 
 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XJX
 
 | | MCU holocomplex in Low-calcium blocking state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
8JSH
 
 | |
6X7K
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8JSG
 
 | |
