8D7K
 
 | |
6CU8
 
 | | Alpha Synuclein fibril formed by full length protein - Twister Polymorph | | Descriptor: | Alpha-synuclein | | Authors: | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | Deposit date: | 2018-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
1BT7
 
 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1XQ8
 
 | |
4A0C
 
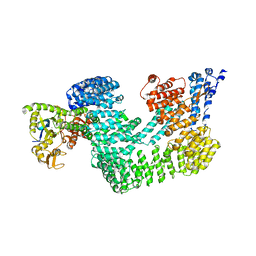 | | Structure of the CAND1-CUL4B-RBX1 complex | | Descriptor: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | Authors: | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
3GAN
 
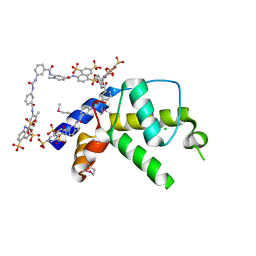 | | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CHLORIDE ION, Uncharacterized protein At3g22680 | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin
To be Published
|
|
1Y6O
 
 | | Crystal structure of disulfide engineered porcine pancreatic phospholipase A2 to group-X isozyme in complex with inhibitor MJ33 and phosphate ions | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
2FUU
 
 | |
6CKA
 
 | | Crystal Structure of Paratox | | Descriptor: | Paratox | | Authors: | Prehna, G. | | Deposit date: | 2018-02-27 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus.
Sci Rep, 8, 2018
|
|
2YB4
 
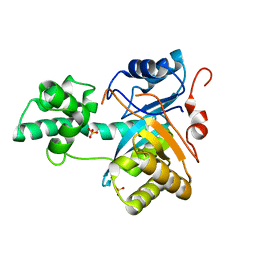 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | Descriptor: | AMIDOHYDROLASE, SULFATE ION | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
1XSM
 
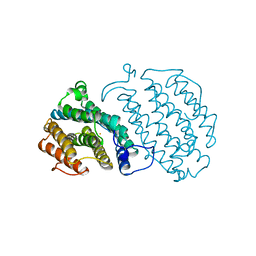 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
1JEQ
 
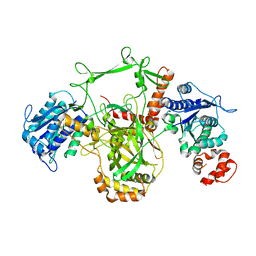 | |
1XNA
 
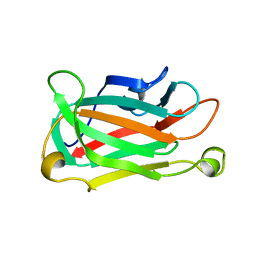 | |
1Y6P
 
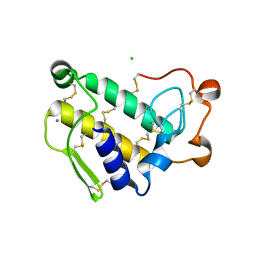 | | Crystal structure of disulfide engineered porcine pancratic phospholipase a2 to group-x isozyme | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
3K4X
 
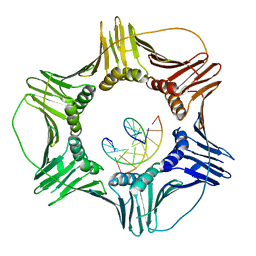 | | Eukaryotic Sliding Clamp PCNA Bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), Proliferating cell nuclear antigen | | Authors: | McNally, R, Kuriyan, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Analysis of the role of PCNA-DNA contacts during clamp loading.
Bmc Struct.Biol., 10, 2010
|
|
4N7G
 
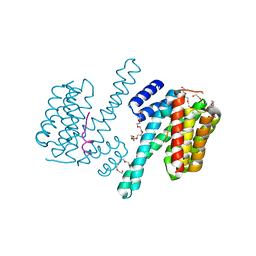 | | Crystal structure of 14-3-3zeta in complex with a peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S, HEXAETHYLENE GLYCOL | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1REP
 
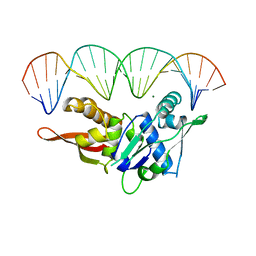 | | CRYSTAL STRUCTURE OF REPLICATION INITIATOR PROTEIN REPE54 OF MINI-F PLASMID COMPLEXED WITH AN ITERON DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Komori, H, Matsunaga, F, Higuchi, Y, Ishiai, M, Wada, C, Miki, K. | | Deposit date: | 1999-04-29 | | Release date: | 2000-02-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 A resolution.
EMBO J., 18, 1999
|
|
2RI7
 
 | |
3R2V
 
 | |
4N84
 
 | | Crystal structure of 14-3-3zeta in complex with a 12-carbon-linker cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1Y6K
 
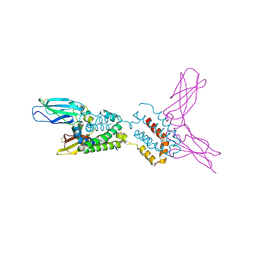 | | Crystal structure of human IL-10 complexed with the soluble IL-10R1 chain | | Descriptor: | Interleukin-10, Interleukin-10 receptor alpha chain | | Authors: | Yoon, S.I, Jones, B.C, Josepson, K, Logsdon, N.J, Walter, M.R. | | Deposit date: | 2004-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain.
Structure, 13, 2005
|
|
2ZOQ
 
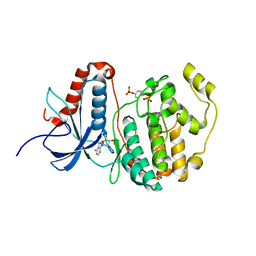 | | Structural dissection of human mitogen-activated kinase ERK1 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | Authors: | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | Deposit date: | 2008-06-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
7CTH
 
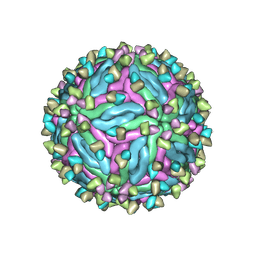 | | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, Single Chain Variable Fragment | | Authors: | Zhang, X, Sharma, A, Duquerroy, S, Zhou, Z.H, Rey, F.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
