4J9C
 
 | |
8BPB
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase core complex at 2.8 Angstrom | | Descriptor: | ACETATE ION, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
8PK2
 
 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PIX
 
 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PJO
 
 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8SH7
 
 | |
4J9G
 
 | |
1JCR
 
 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE NON-SUBSTRATE TETRAPEPTIDE INHIBITOR CVFM AND FARNESYL DIPHOSPHATE SUBSTRATE | | Descriptor: | ACETIC ACID, FARNESYL DIPHOSPHATE, PROTEIN FARNESYLTRANSFERASE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JIE
 
 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with metal-free bleomycin | | Descriptor: | BLEOMYCIN A2, bleomycin-binding protein | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
1JCQ
 
 | | CRYSTAL STRUCTURE OF HUMAN PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYL DIPHOSPHATE AND THE PEPTIDOMIMETIC INHIBITOR L-739,750 | | Descriptor: | 2(S)-{2(S)-[2(R)-AMINO-3-MERCAPTO]PROPYLAMINO-3(S)-METHYL}PENTYLOXY-3-PHENYLPROPIONYLMETHIONINE SULFONE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1QTM
 
 | | DDTTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DT))-3', DNA POLYMERASE I, ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-28 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8BGF
 
 | |
4EO7
 
 | | Crystal structure of the TIR domain of human myeloid differentiation primary response protein 88. | | Descriptor: | MAGNESIUM ION, Myeloid differentiation primary response protein MyD88 | | Authors: | Snyder, G.A, Cirl, C, Jiang, J.S, Chen, P, Smith, T, Xiao, T.S. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8BW1
 
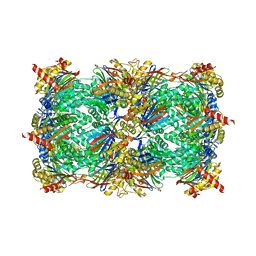 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
4CV9
 
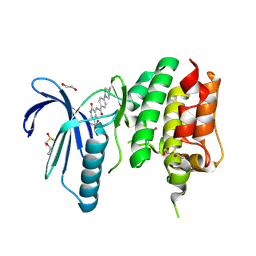 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-Methylpiperazin-1-yl)-N-(2-oxo-5-(pyridin-4-yl)-1,2-dihydropyridin-3-yl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
1CRJ
 
 | |
3ETX
 
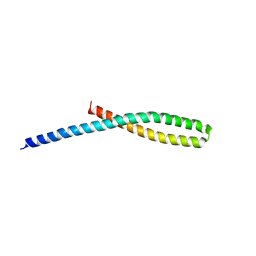 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETZ
 
 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
4JEJ
 
 | | GGGPS from Flavobacterium johnsoniae | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Peterhoff, D, Beer, B, Rajendran, C, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
4CZS
 
 | | Discovery of Glycomimetic Ligands via Genetically-encoded Library of Phage displaying Mannose-peptides | | Descriptor: | 2-hydroxyethyl alpha-D-mannopyranoside, CALCIUM ION, Concanavalin V, ... | | Authors: | Ng, S, Lin, E, Tjhung, K.F, Gerlits, O, Sood, A, Kasper, B, Deng, L, Kitov, P.I, Matochko, W.L, Paschal, B.M, Noren, C.J, Klassen, J, Mahal, L.K, Coates, L, Woods, R.J, Derda, R. | | Deposit date: | 2014-04-22 | | Release date: | 2015-04-22 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Genetically-Encoded Fragment-Based Discovery of Glycopeptide Ligands for Carbohydrate-Binding Proteins.
J.Am.Chem.Soc., 137, 2015
|
|
4JJD
 
 | |
1PHW
 
 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog 1-deoxy-A5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
2QAP
 
 | |
