7A3N
 
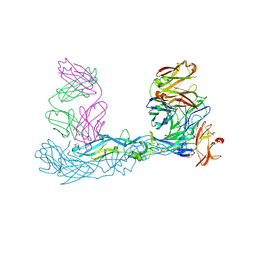 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | CALCIUM ION, Core protein, EDE1 C10 Fab | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3T
 
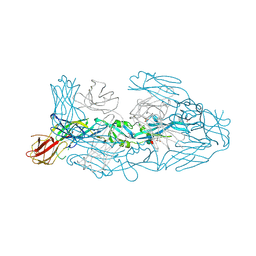 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | Core protein, EDE1 C8 antibody Fab fragment, GLYCEROL, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3U
 
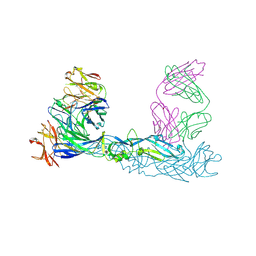 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
4XJE
 
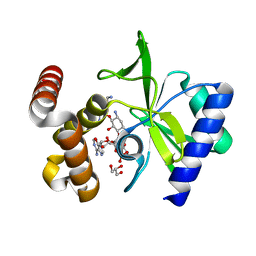 | |
7S68
 
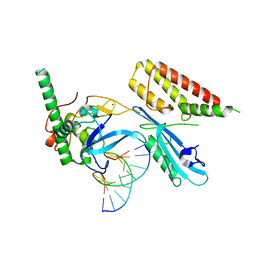 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
6EWW
 
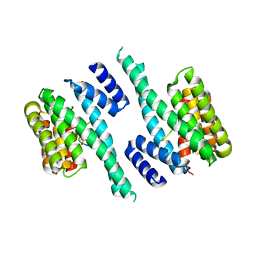 | |
6F08
 
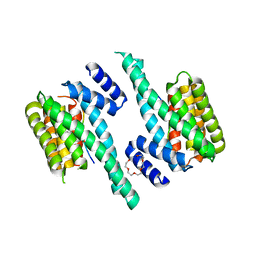 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
7AXZ
 
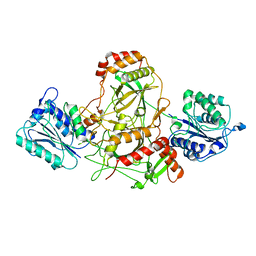 | | Ku70/80 complex apo form | | Descriptor: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | Deposit date: | 2020-11-10 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
7Z13
 
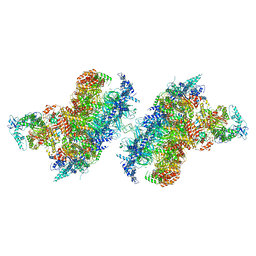 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
1DU0
 
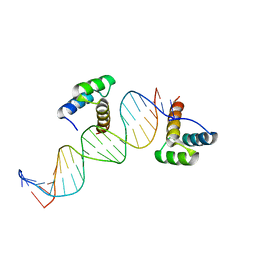 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
5YIU
 
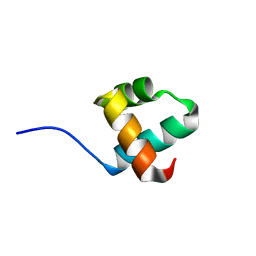 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIW
 
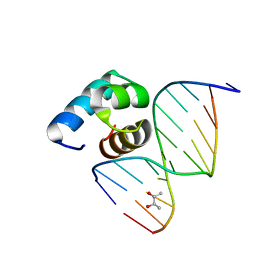 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) in complex with methylated dsDNA (crystal form 2) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*(6MA)P*TP*TP*CP*GP*C)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
3MRJ
 
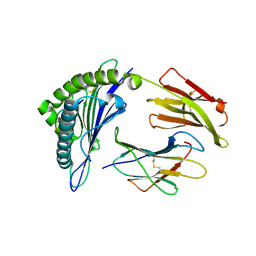 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant
To be Published
|
|
6F09
 
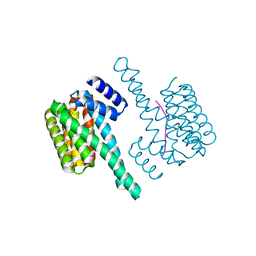 | | Binary complex of 14-3-3 zeta with ubiquitin specific protease 8 (USP8) pSer718 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Ubiquitin carboxyl-terminal hydrolase 8 | | Authors: | Centorrino, F, Ballone, A, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biophysical and structural insight into the USP8/14-3-3 interaction.
FEBS Lett., 592, 2018
|
|
5YIV
 
 | |
3ICH
 
 | |
3ICI
 
 | |
6FNC
 
 | |
6FN9
 
 | |
7DOC
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 5 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Core protein, Genome polyprotein, ... | | Authors: | Quek, J.P. | | Deposit date: | 2020-12-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | 2-Cyanoisonicotinamide Conjugation: A Facile Approach to Generate Potent Peptide Inhibitors of the Zika Virus Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
3IDB
 
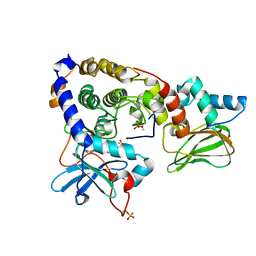 | | Crystal structure of (108-268)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
6YOS
 
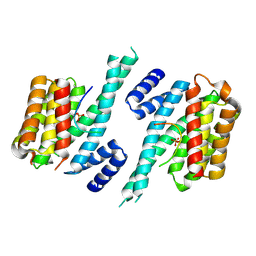 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pT524 pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Glucocorticoid receptor,Glucocorticoid receptor | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
6YMO
 
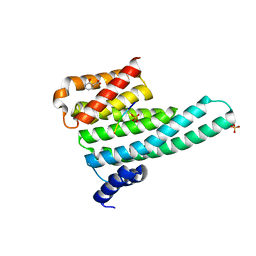 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, 2-HYDROXYBENZOIC ACID, Glucocorticoid receptor, ... | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
6YO8
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pT524 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Glucocorticoid receptor | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
3IDC
 
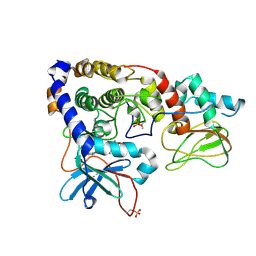 | | Crystal structure of (102-265)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
