2ON9
 
 | |
4IVF
 
 | | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit | | Descriptor: | CITRIC ACID, GLUTATHIONE, Putative uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit
To be Published
|
|
3U40
 
 | | Crystal structure of a purine nucleoside phosphorylase from Entamoeba histolytica bound to adenosine | | Descriptor: | ADENOSINE, NITRATE ION, PHOSPHATE ION, ... | | Authors: | Edwards, T.E, Gardberg, A.S, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-10-06 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression of proteins in Escherichia coli as fusions with maltose-binding protein to rescue non-expressed targets in a high-throughput protein-expression and purification pipeline.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2ONX
 
 | | NNQQ peptide corresponding to residues 8-11 of yeast prion sup35 (alternate crystal form) | | Descriptor: | peptide corresponding to residues 8-11 of yeast prion sup35 | | Authors: | Sawaya, M.R, Sambashivan, S, Nelson, R, Ivanova, M, Sievers, S.A, Apostol, M.I, Thompson, M.J, Balbirnie, M, Wiltzius, J.J, McFarlane, H, Madsen, A.O, Riekel, C, Eisenberg, D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Atomic structures of amyloid cross-beta spines reveal varied steric zippers.
Nature, 447, 2007
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
4ISW
 
 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
4IWH
 
 | |
4F6I
 
 | |
3H53
 
 | | Crystal Structure of human alpha-N-acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminidase, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-04-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
4F6T
 
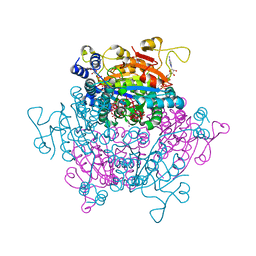 | | The crystal structure of the molybdenum storage protein (MoSto) from Azotobacter vinelandii loaded with various polyoxometalates | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MO(6)-O(26) Cluster, ... | | Authors: | Kowalewski, B, Poppe, J, Schneider, K, Demmer, U, Warkentin, E, Ermler, U. | | Deposit date: | 2012-05-15 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature's Polyoxometalate Chemistry: X-ray Structure of the Mo Storage Protein Loaded with Discrete Polynuclear Mo-O Clusters.
J.Am.Chem.Soc., 134, 2012
|
|
2OOZ
 
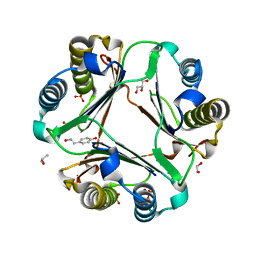 | | Macrophage Migration Inhibitory Factor (MIF) Complexed with OXIM6 (an OXIM Derivative Not Containing a Ring in its R-group) | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(3,3-DIMETHYLBUTANOYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
4IYQ
 
 | |
2H1S
 
 | |
3U8A
 
 | |
2OQ0
 
 | | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | CHLORIDE ION, Gamma-interferon-inducible protein Ifi-16 | | Authors: | Lam, R, Liao, J.C.C, Ravichandran, M, Ma, J, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16
To be Published
|
|
4EZB
 
 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
3U9X
 
 | | Covalent attachment of pyridoxal-phosphate derivatives to 14-3-3 proteins | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-09 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent attachment of pyridoxal-phosphate derivatives to 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3H7P
 
 | | Crystal structure of K63-linked di-ubiquitin | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Weeks, S.D, Grasty, K.C, Hernandez-Cuebas, L, Loll, P.J. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture.
Proteins, 77, 2009
|
|
4F04
 
 | | A Second Allosteric site in E. coli Aspartate Transcarbamoylase: R-state ATCase with UTP bound | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Peterson, A.W, Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A second allosteric site in Escherichia coli aspartate transcarbamoylase.
Biochemistry, 51, 2012
|
|
3GOV
 
 | | Crystal structure of the catalytic region of human MASP-1 | | Descriptor: | GLYCEROL, MASP-1 | | Authors: | Harmat, V, Dobo, J, Beinrohr, L, Sebestyen, E, Zavodszky, P, Gal, P. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MASP-1, a promiscuous complement protease: structure of its catalytic region reveals the basis of its broad specificity.
J.Immunol., 183, 2009
|
|
3TZF
 
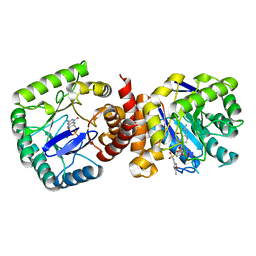 | |
3GL9
 
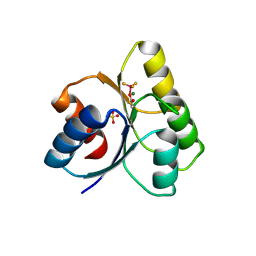 | | The structure of a histidine kinase-response regulator complex sheds light into two-component signaling and reveals a novel cis autophosphorylation mechanism | | Descriptor: | MAGNESIUM ION, Response regulator, SULFATE ION | | Authors: | Casino, P, Rubio, V, Marina, A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction
Cell(Cambridge,Mass.), 139, 2009
|
|
3GLJ
 
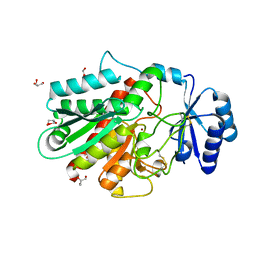 | | A polymorph of carboxypeptidase B zymogen structure | | Descriptor: | Carboxypeptidase B, GLYCEROL, ZINC ION | | Authors: | Fernandez, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Analysis of a new crystal form of procarboxypeptidase B: further insights into the catalytic mechanism
Biopolymers, 2009
|
|
4IYD
 
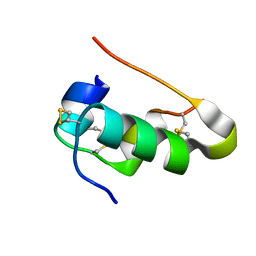 | |
3GQ1
 
 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
