8PDJ
 
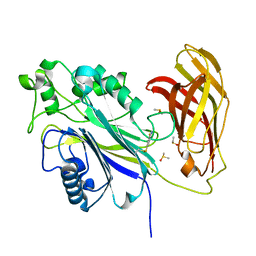 | | The phosphatase and C2 domains of SHIP1 with covalent Z56948267 | | Descriptor: | 4-azanyl-3-fluoranyl-benzenethiol, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8PDH
 
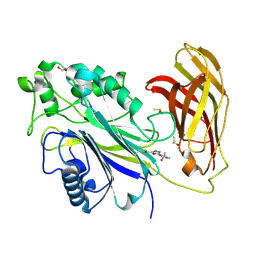 | | The phosphatase and C2 domains of SHIP1 with covalent Z1742148362 | | Descriptor: | (5-phenyl-1,3,4-oxadiazol-2-yl)methanimine, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8PDG
 
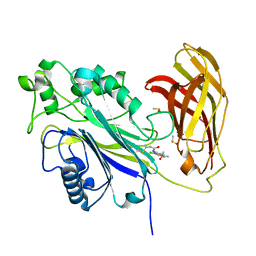 | | The phosphatase and C2 domains of SHIP1 with covalent Z2738285202 | | Descriptor: | DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1, ~{N}-(8-chloranylquinolin-2-yl)propanamide | | Authors: | Bradshaw, W.J, Moreira, T, Scacioc, A, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
1HQA
 
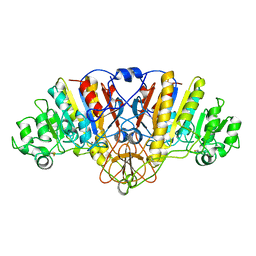 | | ALKALINE PHOSPHATASE (H412Q) | | Descriptor: | ALKALINE PHOSPHATASE, ZINC ION | | Authors: | Ma, L, Kantrowitz, E.R. | | Deposit date: | 1995-11-30 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and X-ray structural studies of a mutant Escherichia coli alkaline phosphatase (His-412-->Gln) at one of the zinc binding sites.
Biochemistry, 35, 1996
|
|
6E6I
 
 | | Crystal structure of 4-methyl HOPDA bound to LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | (1Z,3E)-5-carboxy-3-methyl-5-oxo-1-phenylpenta-1,3-dien-1-olate, 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C, Hurst, T.E, Snieckus, V, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2018-07-24 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal- and Serine-Dependent Meta-Cleavage Product Hydrolases Utilize Similar Nucleophile-Activation Strategies
Acs Catalysis, 8, 2018
|
|
8F56
 
 | |
8F4N
 
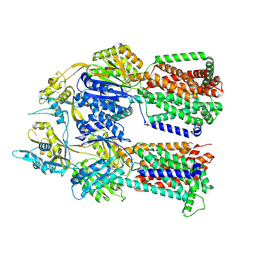 | | Dimer of aminoglycoside efflux pump AcrD | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
8F3E
 
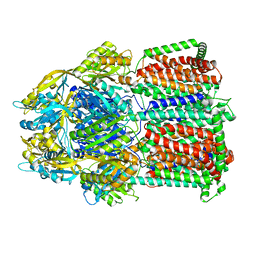 | | Trimer of aminoglycoside efflux pump AcrD | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
4XCT
 
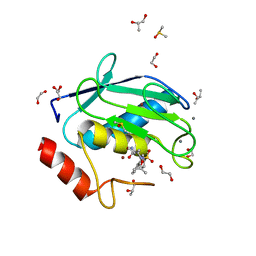 | | Crystal structure of a hydroxamate based inhibitor ARP101 (EN73) in complex with the MMP-9 catalytic domain. | | Descriptor: | (2S,3S)-butane-2,3-diol, (2~{R})-3-methyl-~{N}-oxidanylidene-2-[(4-phenylphenyl)sulfonyl-propan-2-yloxy-amino]butanamide, 1,2-ETHANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Nuti, E, Dive, V, Cassar-Lajeunesse, E, Vera, L, Rossello, A. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
7PP7
 
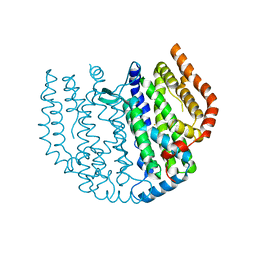 | | Thunberia alata 16:0-ACP desaturase | | Descriptor: | Acyl-[acyl-carrier-protein] 6-desaturase, FE (III) ION | | Authors: | Guy, J.E, Whittle, E, Cai, Y, Chai, J, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2021-09-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselectivity mechanism of the Thunbergia alata Delta 6-16:0-acyl carrier protein desaturase.
Plant Physiol., 188, 2022
|
|
6NZ8
 
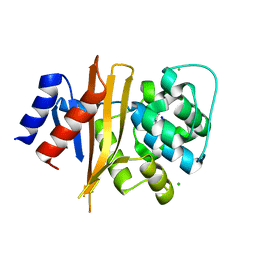 | | Structure of carbamylated apo OXA-231 carbapenemase | | Descriptor: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | Authors: | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
8TB5
 
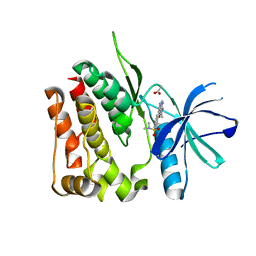 | | TYK2 JH2 bound to Compound7 | | Descriptor: | ACETATE ION, N-{(3P)-3-[3-(dimethylsulfamoyl)phenyl]-1H-pyrrolo[2,3-c]pyridin-5-yl}cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
6O2E
 
 | |
6O2F
 
 | |
8TB6
 
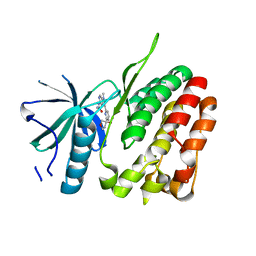 | | TYK2 JH2 bound to Compound14 | | Descriptor: | N-[(3M)-3-{6-[(3R)-3-methoxyoxolan-3-yl]pyridin-2-yl}-1-methyl-1H-pyrrolo[2,3-c]pyridin-5-yl]urea, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
5EAI
 
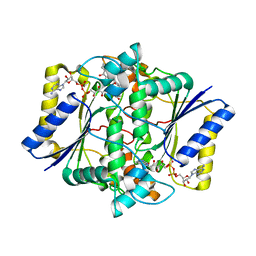 | | Crystal Structure of NAD(P)H dehydrogenase, quinone 1 complexed with a chemotherapeutic naphthoquinone E6a | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{S})-3-bromanyl-1,4-bis(oxidanylidene)-2,3-dihydronaphthalen-2-yl]-3-oxidanyl-2,3-dihydronaphthalene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
5LRV
 
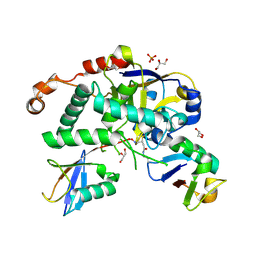 | | Structure of Cezanne/OTUD7B OTU domain bound to Lys11-linked diubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, PHOSPHATE ION, ... | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
7Q28
 
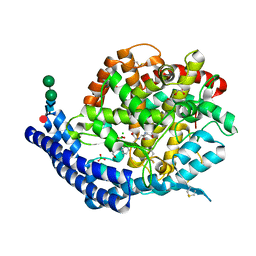 | |
7Q27
 
 | |
7Q29
 
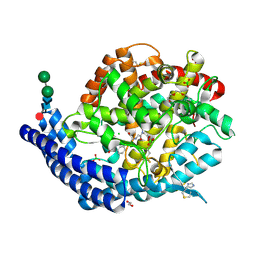 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
7S3F
 
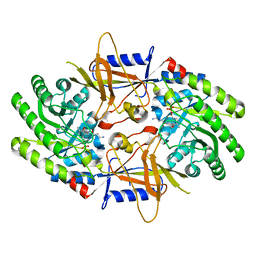 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
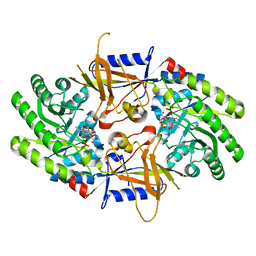 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
6XOZ
 
 | |
6OAV
 
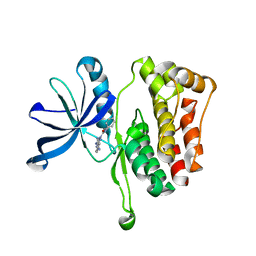 | | JAK2 JH2 in complex with JAK146 | | Descriptor: | 5-amino-3-[(4-cyanophenyl)amino]-N-phenyl-1H-1,2,4-triazole-1-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Puleo, D.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
8EGW
 
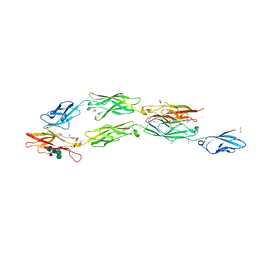 | | Complex of Fat4(EC1-4) bound to Dchs1(EC1-3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Medina, E, Luca, V.C. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the planar cell polarity cadherins Fat4 and Dachsous1.
Nat Commun, 14, 2023
|
|
