8PNV
 
 | | Cryo-EM structure of styrene oxide isomerase | | Descriptor: | Nanobody, PROTOPORPHYRIN IX CONTAINING FE, Styrene oxide isomerase | | Authors: | Khanppnavar, B, Korkhov, B, Li, X. | | Deposit date: | 2023-07-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.048 Å) | | Cite: | Structural basis of the Meinwald rearrangement catalysed by styrene oxide isomerase.
Nat.Chem., 2024
|
|
8PNU
 
 | |
8QZ2
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an inhibitory nanobody (Nb61) | | Descriptor: | Nanobody 61, POTASSIUM ION, Potassium channel subfamily K member 10 | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
2WPM
 
 | | factor IXa superactive mutant, EGR-CMK inhibited | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
8QZ3
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ4
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb76) | | Descriptor: | BARIUM ION, CHOLESTEROL HEMISUCCINATE, Nanobody 76, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
8STL
 
 | | Crystal Structure of Nanobody PIK3_Nb16 against wild-type PI3Kalpha | | Descriptor: | Nanobody PIK3_Nb16, SULFATE ION | | Authors: | Nwafor, J.N, Srinivasan, L, Chen, Z, Gabelli, S.B, Iheanacho, A, Alzogaray, V, Klinke, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of isoform specific nanobodies for Class I PI3Ks
To be published
|
|
8VT2
 
 | | cryo-EM structure of HMPV (MPV-2c) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prefusion of HMPV (MPV-2c), ... | | Authors: | Yu, X, Langedijk, J.P.M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer.
Nat Commun, 15, 2024
|
|
8VT3
 
 | | cryo-EM structure of HMPV (MPV-2cREKR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prefusion of HMPV (MPV-2cREKR), ... | | Authors: | Yu, X, Langedijk, J.P.M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer.
Nat Commun, 15, 2024
|
|
2WPH
 
 | | factor IXa superactive triple mutant | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPK
 
 | | factor IXa superactive triple mutant, ethylene glycol-soaked | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2XAC
 
 | |
2WPL
 
 | | factor IXa superactive triple mutant, EDTA-soaked | | Descriptor: | COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, D-PHE-PRO-ARG-CHLOROMETHYL KETONE | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPJ
 
 | | factor IXa superactive triple mutant, NaCl-soaked | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2XA8
 
 | | Crystal structure of the Fab domain of omalizumab at 2.41A | | Descriptor: | OMALIZUMAB HEAVY CHAIN, OMALIZUMAB LIGHT CHAIN | | Authors: | Huang, C.H, Hung, F.H.A, Lim, C, Chang, T.W, Ma, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Physical Basis for Anti-IgE Therapy.
Sci Rep, 5, 2015
|
|
2XBO
 
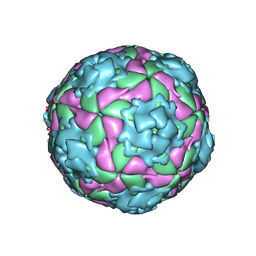 | | Equine Rhinitis A Virus in Complex with its Sialic Acid Receptor | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, P1 | | Authors: | Fry, E.E, Tuthill, T.J, Harlos, K, Walter, T.S, Rowlands, D.J, Stuart, D.I. | | Deposit date: | 2010-04-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of Equine Rhinitis a Virus in Complex with its Sialic Acid Receptor.
J.Gen.Virol., 91, 2010
|
|
2XSC
 
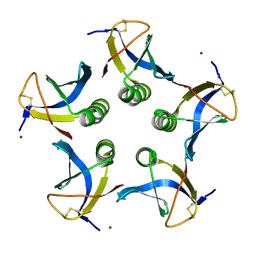 | | Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli | | Descriptor: | SHIGA-LIKE TOXIN 1 SUBUNIT B, ZINC ION | | Authors: | Stein, P.E, Boodhoo, A, Tyrrell, G.J, Brunton, J.L, Oeffner, R.D, Bunkoczi, G, Read, R.J. | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of the Cell-Binding B Oligomer of Verotoxin-1 from E. Coli.
Nature, 355, 1992
|
|
4X7P
 
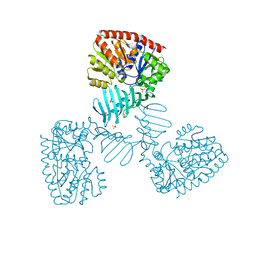 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3LE2
 
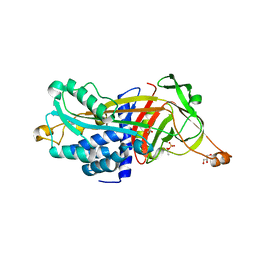 | | Structure of Arabidopsis AtSerpin1. Native Stressed Conformation | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Harrop, S.J, Joss, T.V, Cumi, P.M.G, Roberts, T.H. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21).
J.Biol.Chem., 285, 2010
|
|
8Y3P
 
 | | ASFV p72 in complex with Fab C9 | | Descriptor: | B646L, Heavy chain of C9, Light chain of C9 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3Q
 
 | | ASFV p72 in complex with Fab F11 | | Descriptor: | B646L, Heavy chain of F11, Light chain of F11 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8V7X
 
 | |
8Y3O
 
 | | ASFV p72 in complex with Fab B1 | | Descriptor: | B646L, Heavy chain of B1, Light chain of B1 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
