6D5A
 
 | |
9EUD
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23c | | Descriptor: | (1-propan-2-ylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
9EU6
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23j | | Descriptor: | (1,5-dimethylpyrazol-4-yl)methyl (2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
8WSS
 
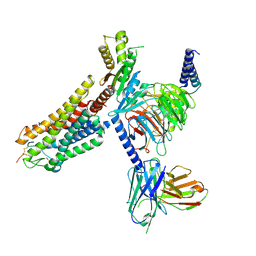 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
8ZFQ
 
 | |
9BKK
 
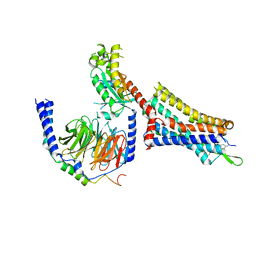 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
6UJW
 
 | | P-glycoprotein mutant-Y306A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
1SLF
 
 | |
9EUB
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 24e | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [1-(2-hydroxyethyl)pyrazol-4-yl]methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
1SLG
 
 | | STREPTAVIDIN, PH 5.6, BOUND TO PEPTIDE FCHPQNT | | Descriptor: | FCHPQNT, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1995-03-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence
Biochemistry, 34, 1995
|
|
9EU8
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15h | | Descriptor: | (4-methyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
2XJK
 
 | | Monomeric Human Cu,Zn Superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Saraboji, K, Leinartaite, L, Nordlund, A, Oliveberg, M, Logan, D.T. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Folding Catalysis by Transient Coordination of Zn2+ to the Cu Ligands of the Als-Associated Enzyme Cu/Zn Superoxide Dismutase 1.
J.Am.Chem.Soc., 132, 2010
|
|
1SLE
 
 | | STREPTAVIDIN, PH 5.0, BOUND TO CYCLIC PEPTIDE AC-CHPQGPPC-NH2 | | Descriptor: | AC-CHPQGPPC-NH2, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1995-03-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence
Biochemistry, 34, 1995
|
|
4GBA
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | DCN1-like protein 3, NEDD8-conjugating enzyme UBE2F | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
9BKJ
 
 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
8ZFN
 
 | |
9AXG
 
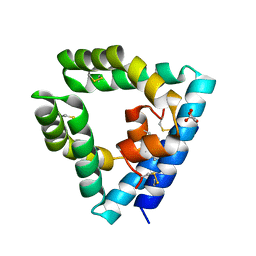 | |
1DEY
 
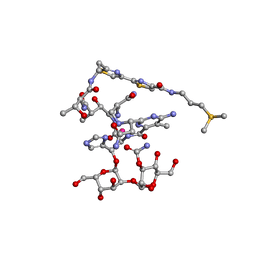 | |
5LKJ
 
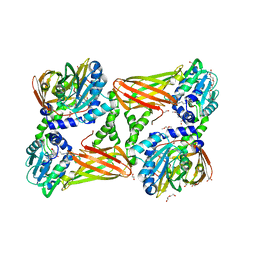 | | Crystal structure of mouse CARM1 in complex with ligand SA684 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-carbamimidamidoethyl)amino]-2-azanyl-butanoic acid, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with ligands
To Be Published
|
|
6ME2
 
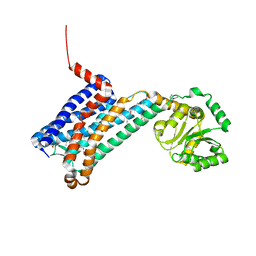 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
3U4N
 
 | | A novel covalently linked insulin dimer | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Norrman, M, Vinther, T.N. | | Deposit date: | 2011-10-10 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel covalently linked insulin dimer engineered to investigate the function of insulin dimerization.
Plos One, 7, 2012
|
|
6ME4
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
3KVW
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
6UIQ
 
 | |
