7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
5VPX
 
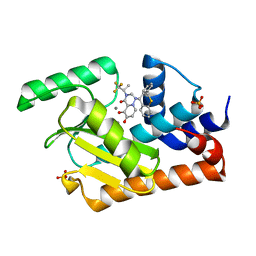 | | I38T mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
5VQN
 
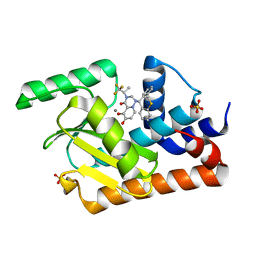 | | E119D mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
5VPT
 
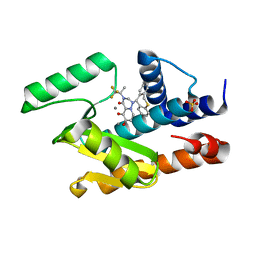 | | 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
7QWS
 
 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
8SCB
 
 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
7TOQ
 
 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein poly-PR | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
7OW7
 
 | |
9EQJ
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-39 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[cis-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9EQM
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
5BTH
 
 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6X
 
 | |
5VY2
 
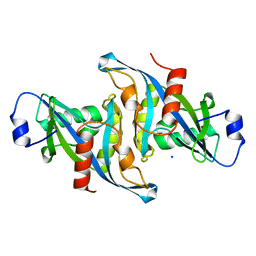 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
6R3V
 
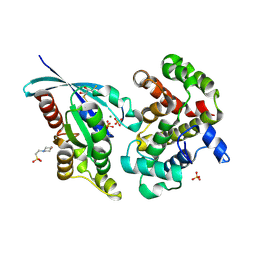 | | Crystal Structure of RhoA-GDP-Pi in Complex with RhoGAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jin, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A GAP-GTPase-GDP-PiIntermediate Crystal Structure Analyzed by DFT Shows GTP Hydrolysis Involves Serial Proton Transfers.
Chemistry, 25, 2019
|
|
7D63
 
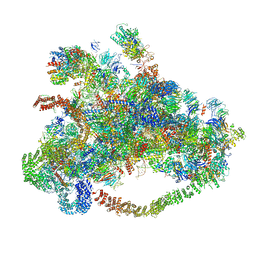 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7WTW
 
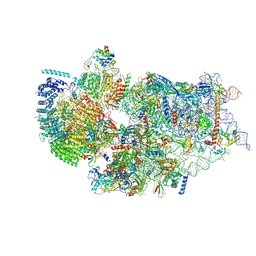 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A3 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
6R1T
 
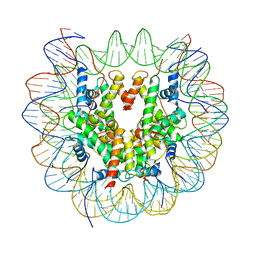 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
7D5S
 
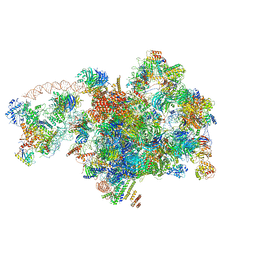 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
2XYA
 
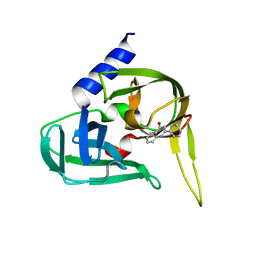 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4O18
 
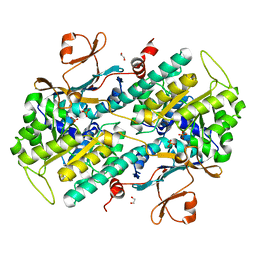 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O17
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1A
 
 | | The crystal structure of the mutant NAMPT G217R | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
