5W4T
 
 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
4K9V
 
 | | Complex of CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(3S,6S)-6-{[N-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-L-seryl]amino}octan-3-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
4K9T
 
 | | Complex of CYP3A4 with a desoxyritonavir analog | | Descriptor: | Cytochrome P450 3A4, N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-(4-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}butyl)-L-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
1IOX
 
 | | NMR Structure of human Betacellulin-2 | | Descriptor: | Betacellulin | | Authors: | Miura, K, Doura, H, Aizawa, T, Tada, H, Seno, M, Yamada, H, Kawano, K. | | Deposit date: | 2001-04-18 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of betacellulin, a new member of EGF-family ligands.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
2RL9
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 6.5 bound to trimannoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Hindsgaul, O, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2007-10-18 | | Release date: | 2008-02-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of pH-dependent ligand binding and release by the cation-dependent mannose 6-phosphate receptor.
J.Biol.Chem., 283, 2008
|
|
6EMN
 
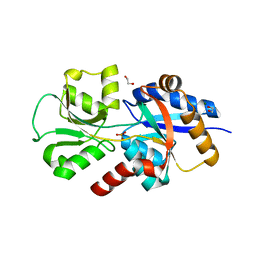 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6V2E
 
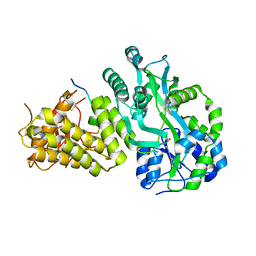 | |
5MNH
 
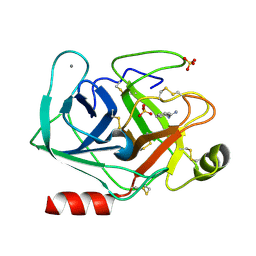 | | Cationic trypsin in complex with benzamidine (deuterated sample at 295 K) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MNO
 
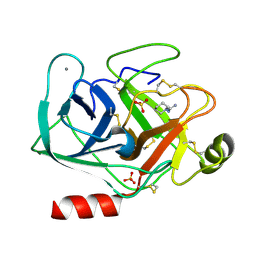 | | Cationic trypsin in complex with N-amidinopiperidine (deuterated sample at 295 K) | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MOQ
 
 | | Joint X-ray/neutron structure of cationic trypsin in complex with benzamidine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (0.93 Å), X-RAY DIFFRACTION | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MOP
 
 | | Joint X-ray/neutron structure of cationic trypsin in its apo form | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (0.99 Å), X-RAY DIFFRACTION | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
5MNF
 
 | |
6Y4M
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb111 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, (2~{S},4~{R})-4-azanyl-2-methyl-5-phenyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
6Y4N
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb116 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
2VFD
 
 | | Crystal structure of the F96S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5MNE
 
 | |
7ZNT
 
 | | CRYSTAL STRUCTURE OF AT7 IN COMPLEX WITH THE SECOND BROMODOMAIN OF HUMAN BRD4 AND PVHL:ELONGINC:ELONGINB | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-[6-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]hexylsulfanyl]-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Hughes, S.J, Casement, R, Ciulli, A. | | Deposit date: | 2022-04-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders.
Nat.Chem.Biol., 19, 2023
|
|
5MNN
 
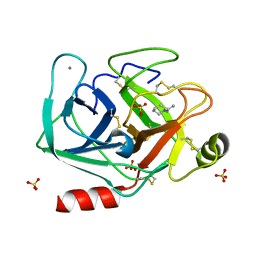 | | Cationic trypsin in complex with N-amidinopiperidine (deuterated sample at 100 K) | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.859 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MO2
 
 | | Neutron structure of cationic trypsin in complex with N-amidinopiperidine | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
1JGN
 
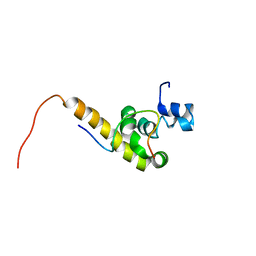 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
3WHC
 
 | |
2W8F
 
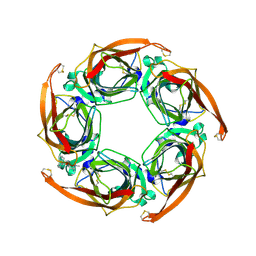 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
6GHQ
 
 | | HtxB D206N protein variant from Pseudomonas stutzeri in a partially open conformation to 1.53 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
5MNQ
 
 | | Cationic trypsin in complex with a derivative of N-amidinopiperidine | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(1-carbamimidoylpiperidin-4-yl)methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
