2QUW
 
 | |
8PK2
 
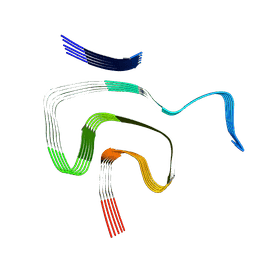 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PIX
 
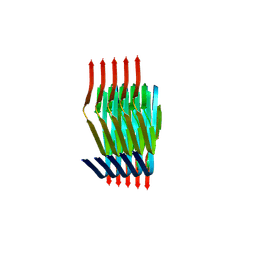 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PJO
 
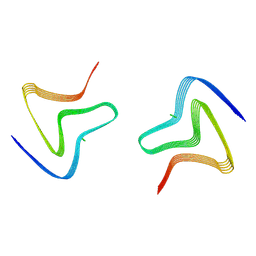 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
4IJJ
 
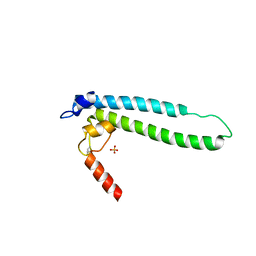 | | Structure of transcription factor DksA2 from Pseudomonas aeruginosa | | Descriptor: | Putative C4-type zinc finger protein, DksA/TraR family, SULFATE ION | | Authors: | Biswas, T, Furman, R, Artsimovitch, I, Tsodikov, O.V. | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-27 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DksA2, a zinc-independent structural analog of the transcription factor DksA.
Febs Lett., 587, 2013
|
|
4IMY
 
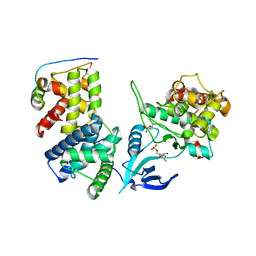 | |
6QHG
 
 | | Structure of the cap-binding domain of Rift Valley Fever virus L protein | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Gogrefe, N, Reindl, S, Gunther, S, Rosenthal, M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure of a functional cap-binding domain in Rift Valley fever virus L protein.
Plos Pathog., 15, 2019
|
|
2I53
 
 | | Crystal structure of Cyclin K | | Descriptor: | ACETATE ION, Cyclin K | | Authors: | Baek, K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2006-08-23 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Cyclin K, a Positive Regulator of Cyclin-dependent Kinase 9.
J.Mol.Biol., 366, 2007
|
|
8P7G
 
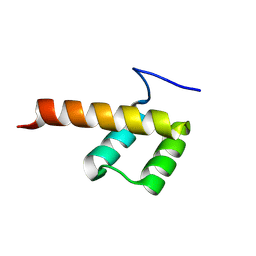 | |
4D0N
 
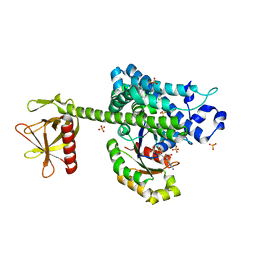 | | AKAP13 (AKAP-Lbc) RhoGEF domain in complex with RhoA | | Descriptor: | 1,2-ETHANEDIOL, A-KINASE ANCHOR PROTEIN 13, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Abdul Azeez, K.R, Shrestha, L, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Klussmann, E, Elkins, J.M. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Rhoa : Akap-Lbc Dh-Ph Domain Complex.
Biochem.J., 464, 2014
|
|
2K0N
 
 | |
8UNJ
 
 | | Atomic model of the human CTF18-RFC alone in the apo state (State 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6GVA
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR4455 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-azanylethylsulfanyl)-3-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, BROMIDE ION, ... | | Authors: | Skerlova, J, Rezacova, P. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d]pyrimidine Inhibitors of Cyclin-Dependent Kinases and Their Evaluation in Lymphoma Models.
J.Med.Chem., 62, 2019
|
|
8QU4
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
1RMU
 
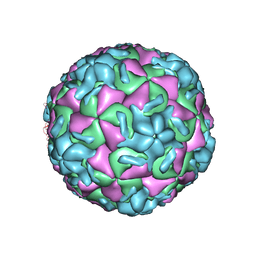 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
1F6M
 
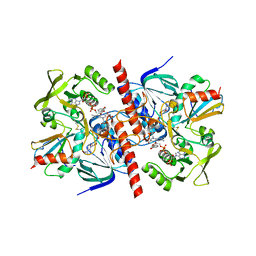 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
3PT2
 
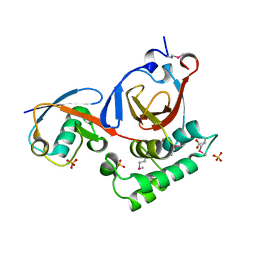 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TN8
 
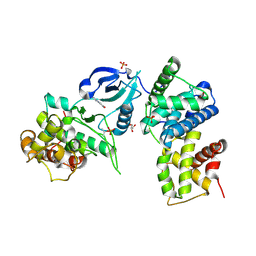 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.E. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3PY1
 
 | | CDK2 ternary complex with SU9516 and ANS | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3TNW
 
 | | Structure of CDK2/cyclin A in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
2LPB
 
 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
3VJT
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(R)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
3VRT
 
 | | VDR ligand binding domain in complex with 2-Mehylidene-19,25,26,27-tetranor-1alpha,24-dihydroxyvitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxypentan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yoshimoto, N, Inaba, Y, Itoh, T, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VJS
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
