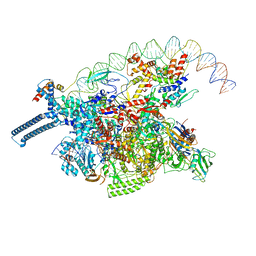
8DY7
 
 | |
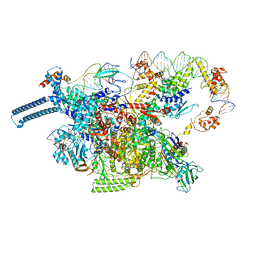
8DY9
 
 | |
7ZF2
 
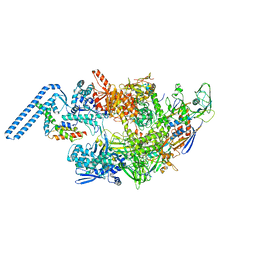 | | Protomeric substructure from an octameric assembly of M. tuberculosis RNA polymerase in complex with sigma-b initiation factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Trapani, S, Bron, P, Lai Kee Him, J, Brodolin, K, Morichaud, Z, Vishwakarma, R. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Nat Commun, 14, 2023
|
|
3DXJ
 
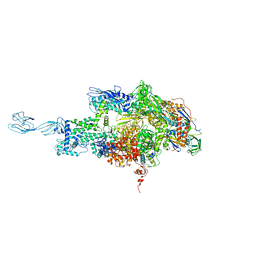 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
5H6X
 
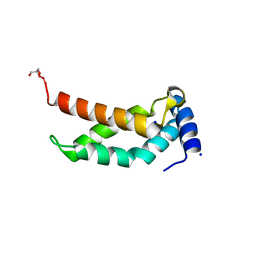 | | The crystal structure of RpoS fragment including a partial region 1.2 and region 2 from intracellular pathogen Legionella pneumophila | | Descriptor: | GLYCEROL, RNA polymerase sigma factor RpoS, SODIUM ION | | Authors: | Zhang, N, Chen, X, Gong, X, Ge, H. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of RpoS Fragment including a Partial Region 1.2 and Region 2 from the Intracellular Pathogen Legionella pneumophila
Crystals, 8, 2018
|
|
3EQL
 
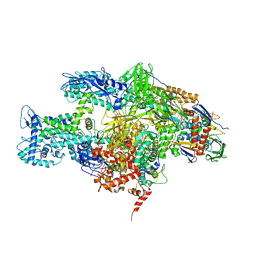 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transcription inactivation through local refolding of the RNA polymerase structure.
Nature, 457, 2009
|
|
4OIR
 
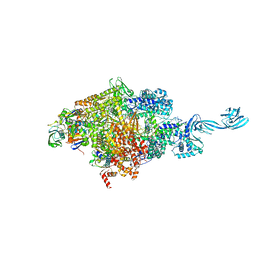 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
8HIH
 
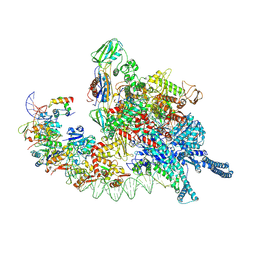 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4OIN
 
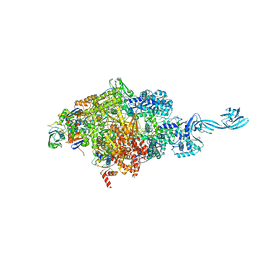 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIO
 
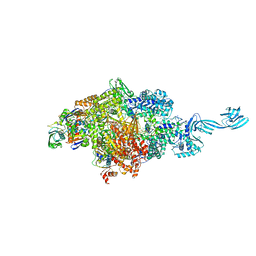 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6CUX
 
 | |
8IGR
 
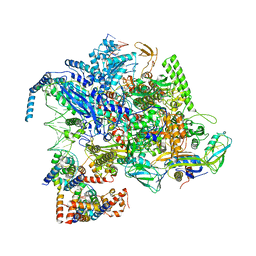 | | Cryo-EM structure of CII-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8IGS
 
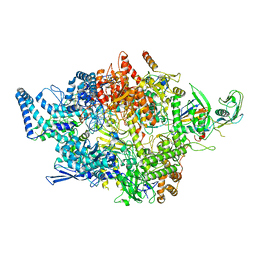 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
6CUU
 
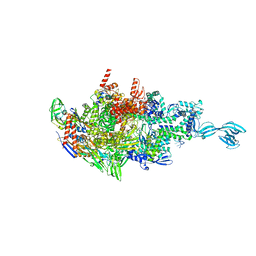 | | Thermus thermophiles RNA polymerase in complex with promoter DNA and antibiotic Kanglemycin A | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*CP*TP*CP*TP*GP*AP*TP*GP*CP*A)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*CP*AP*GP*AP*GP*CP*CP*CP*AP*AP*AP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Mode of Action of Kanglemycin A, an Ansamycin Natural Product that Is Active against Rifampicin-Resistant Mycobacterium tuberculosis.
Mol. Cell, 72, 2018
|
|
6DCF
 
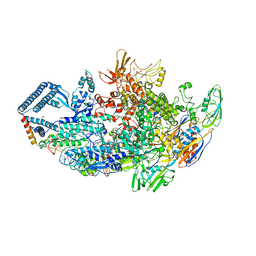 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
7Q59
 
 | |
7RDQ
 
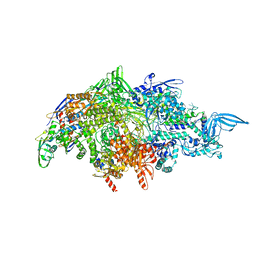 | |
1L9U
 
 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
1KU2
 
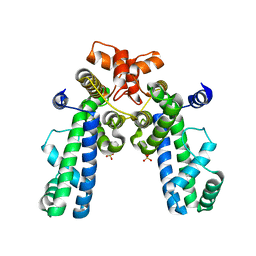 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment Containing Regions 1.2 to 3.1 | | Descriptor: | SULFATE ION, sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
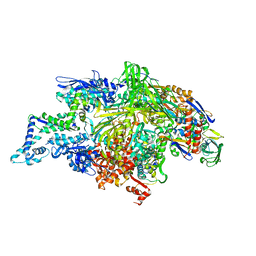
7Q4U
 
 | |
1L9Z
 
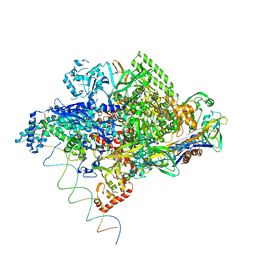 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
4ZH3
 
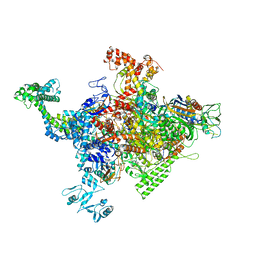 | |
5VO8
 
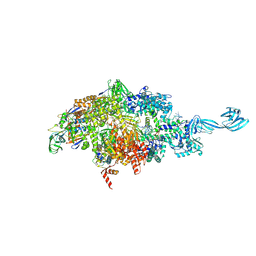 | | X-ray crystal structure of a bacterial reiterative transcription complex of pyrG promoter | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*CP*TP*GP*AP*TP*GP*CP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*GP*CP*AP*TP*CP*AP*GP*AP*GP*CP*CP*CP*AP*AP*AP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Murakami, K.S, Shin, Y, Turnbough Jr, C.L, Molodtsov, V. | | Deposit date: | 2017-05-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VT0
 
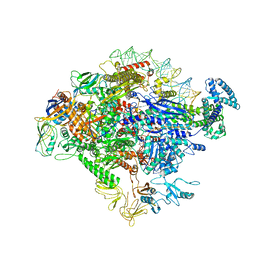 | |
5VI5
 
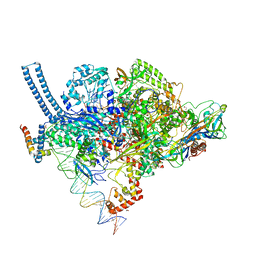 | | Structure of Mycobacterium smegmatis transcription initiation complex with a full transcription bubble | | Descriptor: | 1,2-ETHANEDIOL, DNA (44-MER), DNA (49-MER), ... | | Authors: | Darst, S.A, Campbell, E.A, Lilic, M. | | Deposit date: | 2017-04-14 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures.
Nat Commun, 8, 2017
|
|
