1PF7
 
 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH IMMUCILLIN H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | De Azevedo Jr, W.F, Canduri, F, Dos Santos, D.M, Pereira, J.H, Dias, M.V.B, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-05-24 | | Release date: | 2004-06-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for inhibition of human PNP by immucillin-H
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1PMN
 
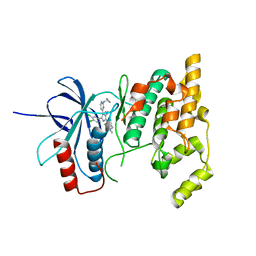 | | Crystal structure of JNK3 in complex with an imidazole-pyrimidine inhibitor | | Descriptor: | CYCLOPROPYL-{4-[5-(3,4-DICHLOROPHENYL)-2-[(1-METHYL)-PIPERIDIN]-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PK7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with adenosine and sulfate/phosphate | | Descriptor: | ADENOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PK9
 
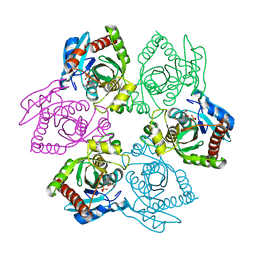 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
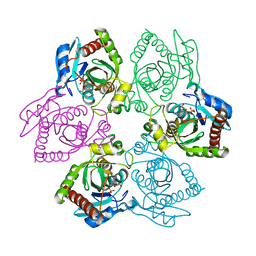 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR6
 
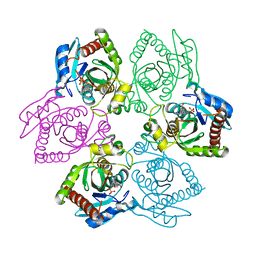 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-xylofuranosyladenine and Phosphate/Sulfate | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1N8Z
 
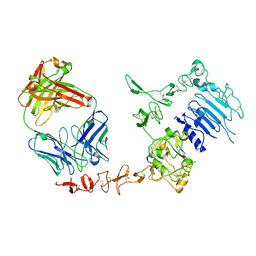 | | Crystal structure of extracellular domain of human HER2 complexed with Herceptin Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Herceptin Fab (antibody) - heavy chain, Herceptin Fab (antibody) - light chain, ... | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in Complex with the Herceptin Fab
Nature, 421, 2003
|
|
1NH3
 
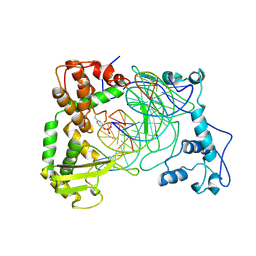 | | Human Topoisomerase I Ara-C Complex | | Descriptor: | 5'-D(*(GNG)P*GP*AP*AP*AP*AP*AP*UP*UP*UP*UP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*UP*(UBB))-3', 5'-D(*AP*AP*AP*AP*AP*TP*UP*UP*UP*UP*CP*(CAR)P*AP*AP*GP*UP*CP*UP*UP*UP*UP*T)-3', ... | | Authors: | Chrencik, J.E, Burgin, A.B, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Impact of the Leukemia Drug 1-beta-D-Arabinofuranosylcytosine (Ara-C) on the Covalent Human Topoisomerase I-DNA Complex
J.Biol.Chem., 278, 2003
|
|
1OKZ
 
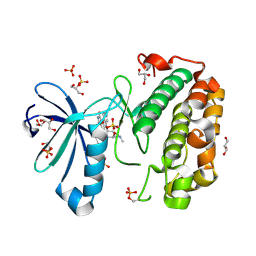 | | Structure of human PDK1 kinase domain in complex with UCN-01 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 7-HYDROXYSTAUROSPORINE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
1OKY
 
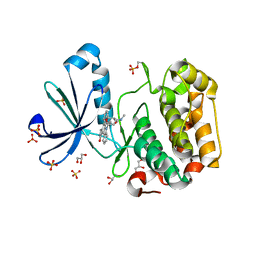 | | Structure of human PDK1 kinase domain in complex with staurosporine | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, STAUROSPORINE, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
1OGK
 
 | | The crystal structure of Trypanosoma cruzi dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE, DEOXYURIDINE-5'-DIPHOSPHATE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1OGL
 
 | | The crystal structure of native Trypanosoma cruzi dUTPase | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1PMV
 
 | | The structure of JNK3 in complex with a dihydroanthrapyrazole inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PYX
 
 | | GSK-3 Beta complexed with AMP-PNP | | Descriptor: | Glycogen synthase kinase-3 beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors
J.Mol.Biol., 333, 2003
|
|
1PR5
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 7-deazaadenosine and Phosphate/Sulfate | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1O80
 
 | |
1OAJ
 
 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castelli, S, Falconi, M, Parrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-27 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
1OC0
 
 | | plasminogen activator inhibitor-1 complex with somatomedin B domain of vitronectin | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1, VITRONECTIN | | Authors: | Read, R.J, Zhou, A, Huntington, J.A, Pannu, N.S, Carrell, R.W. | | Deposit date: | 2003-02-03 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | How Vitronectin Binds Pai-1 to Modulate Fibrinolysis and Cell Migration
Nat.Struct.Biol., 10, 2003
|
|
1P42
 
 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P19
 
 | |
1OM1
 
 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
1OSV
 
 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1P4Y
 
 | |
1PJZ
 
 | |
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
