8X7A
 
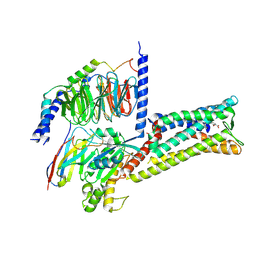 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
6HUS
 
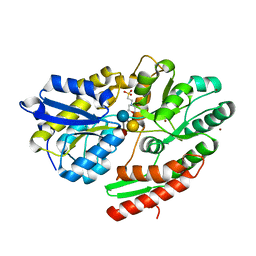 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
1S4I
 
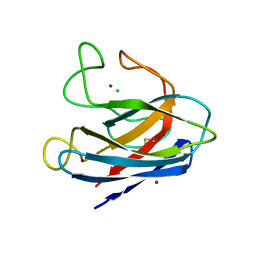 | | Crystal structure of a SOD-like protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, ZINC ION, superoxide dismutase-like protein yojM | | Authors: | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | Deposit date: | 2004-01-16 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8I54
 
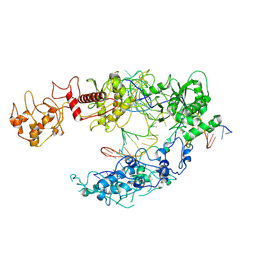 | | Lb2Cas12a RNA DNA complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3'), Lb2Cas12a, ... | | Authors: | Li, J, Sivaraman, J, Satoru, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
8V9I
 
 | | 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Deinococcus radiodurans with D-phenylalanine-derived triazole acetylphosphonate (D-PheTrAP) bound | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, 2-HYDROXY BUTANE-1,4-DIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(1S)-1-[(S)-(2-{1-[(1R)-1-carboxy-2-phenylethyl]-1H-1,2,3-triazol-4-yl}ethoxy)(hydroxy)phosphoryl]-1-hydroxyethyl}-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Potent Inhibition of E. coli DXP Synthase by a gem -Diaryl Bisubstrate Analog.
Acs Infect Dis., 10, 2024
|
|
8UKE
 
 | |
8V0P
 
 | |
8UK1
 
 | |
8V0T
 
 | |
8V0O
 
 | |
8V0M
 
 | |
6HUR
 
 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 2'-fucosyllactose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Katayama, T, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
7RAN
 
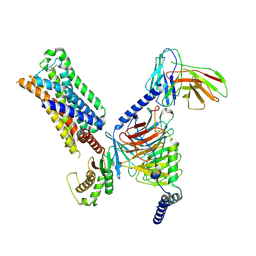 | | 5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (3R)-3-methyl-5-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,3,6-tetrahydropyridin-1-ium, 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Nature, 610, 2022
|
|
3VYR
 
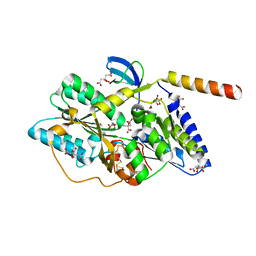 | | Crystal structure of the HypC-HypD complex | | Descriptor: | CITRIC ACID, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYS
 
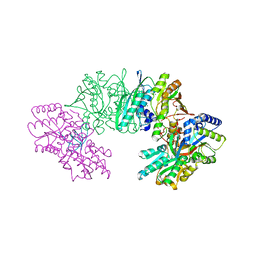 | | Crystal structure of the HypC-HypD-HypE complex (form I) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
7RB3
 
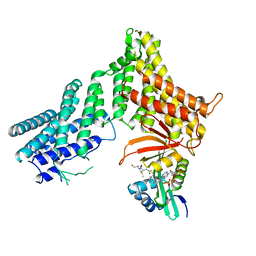 | |
8SMV
 
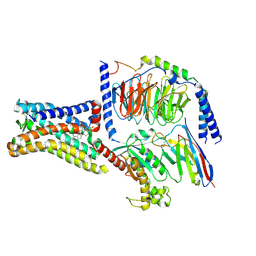 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
7RAP
 
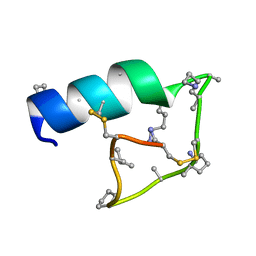 | |
6IYK
 
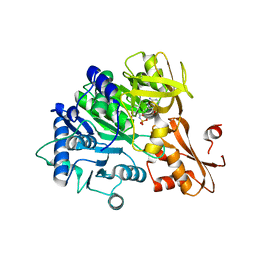 | | The structure of EntE with 2-nitrobenzoyl adenylate analog | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(2-nitrobenzene-1-carbonyl)sulfamoyl]adenosine | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6YV2
 
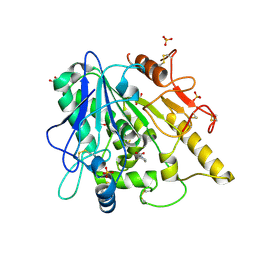 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | Descriptor: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
8U1D
 
 | | Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1285 Heavy Chain, DH1285 Light Chain, ... | | Authors: | Thakur, B, Stalls, V.D, Acharya, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques.
Cell, 187, 2024
|
|
2VSU
 
 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
8V0L
 
 | |
2ICT
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
8UIR
 
 | |
