2B7L
 
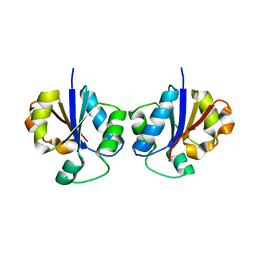 | | Crystal Structure of CTP:Glycerol-3-Phosphate Cytidylyltransferase from Staphylococcus aureus | | Descriptor: | glycerol-3-phosphate cytidylyltransferase | | Authors: | Fong, D.H, Yim, V.C.-N, D'Elia, M.A, Brown, E.D, Berghuis, A.M. | | Deposit date: | 2005-10-04 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of CTP:glycerol-3-phosphate cytidylyltransferase from Staphylococcus aureus: examination of structural basis for kinetic mechanism.
Biochim.Biophys.Acta, 1764, 2006
|
|
5LM5
 
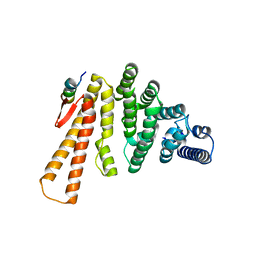 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM2 peptide (region 435-451) | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, mRNA decapping protein 2 | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMG
 
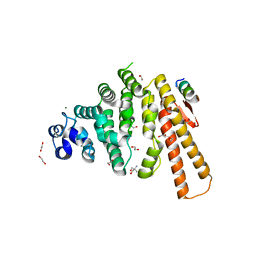 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM10 peptide (region 954-970) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LQG
 
 | |
3BP2
 
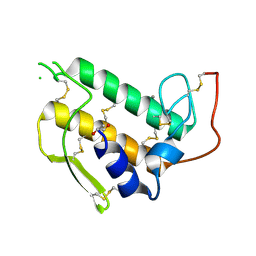 | |
3LPO
 
 | | Crystal structure of the N-terminal domain of sucrase-isomaltase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sucrase-isomaltase, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains.
J.Biol.Chem., 285, 2010
|
|
3LVR
 
 | | The crystal structure of ASAP3 in complex with Arf6 in transition state soaked with Calcium | | Descriptor: | ALUMINUM FLUORIDE, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 3, ... | | Authors: | Ismail, S.A, Vetter, I.R, Sot, B, Wittinghofer, A. | | Deposit date: | 2010-02-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism
Cell(Cambridge,Mass.), 141, 2010
|
|
3LSE
 
 | | N-Domain of human adhesion/growth-regulatory galectin-9 in complex with lactose | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
3LUE
 
 | | Model of alpha-actinin CH1 bound to F-actin | | Descriptor: | Actin, cytoplasmic 1, Alpha-actinin-3 | | Authors: | Galkin, V.E, Orlova, A, Salmazo, A, Djinovic-Carugo, K, Egelman, E.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Opening of tandem calponin homology domains regulates their affinity for F-actin.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LZK
 
 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3LZY
 
 | |
3M61
 
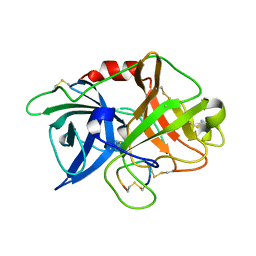 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
3M7Q
 
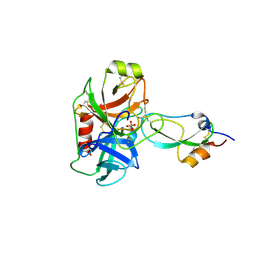 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
3M4O
 
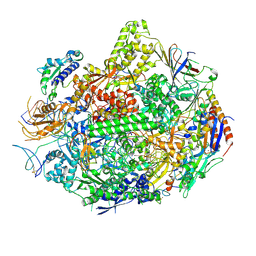 | | RNA polymerase II elongation complex B | | Descriptor: | DNA (28-MER), DNA (5'-D(P*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-11 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MSL
 
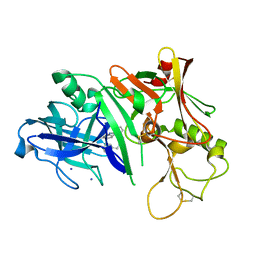 | |
3MA2
 
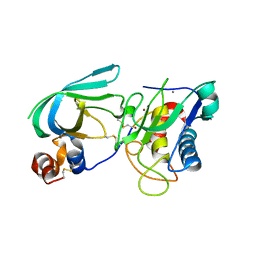 | | Complex membrane type-1 matrix metalloproteinase (MT1-MMP) with tissue inhibitor of metalloproteinase-1 (TIMP-1) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-14, Metalloproteinase inhibitor 1, ... | | Authors: | Grossman, M, Tworowski, D, Dym, O, Lee, M.-H, Levy, Y, Sagi, I. | | Deposit date: | 2010-03-23 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Intrinsic Protein Flexibility of Endogenous Protease Inhibitor TIMP-1 Controls Its Binding Interface and Affects Its Function.
Biochemistry, 49, 2010
|
|
3MSK
 
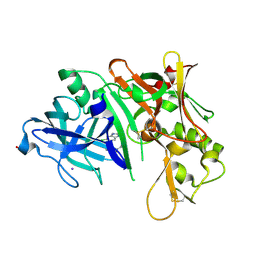 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | Descriptor: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MHZ
 
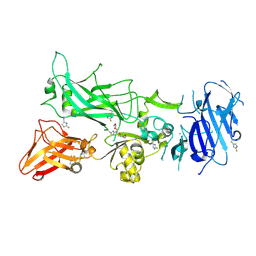 | | 1.7A structure of 2-fluorohistidine labeled Protective Antigen | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Wimalasena, D.S, Janowiak, B.E, Miyagi, M, Sun, J, Hajduch, J, Pooput, C, Kirk, K.L, Bann, J.G. | | Deposit date: | 2010-04-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry, 49, 2010
|
|
3LKU
 
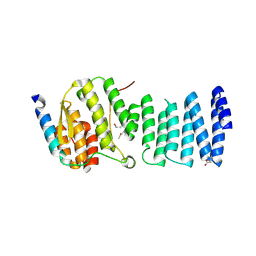 | |
4TX4
 
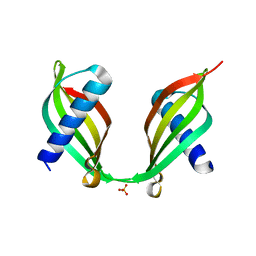 | | Crystal Structure of a Single-Domain Cysteine Protease Inhibitor from Cowpea (Vigna unguiculata) | | Descriptor: | Cysteine proteinase inhibitor, SULFATE ION | | Authors: | Pereira, H.M, Valadares, N, Monteiro-Junior, J.E, Carvalho, C.P.S, Grangeiro, T.B. | | Deposit date: | 2014-07-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression in Escherichia coli of cysteine protease inhibitors from cowpea (Vigna unguiculata): The crystal structure of a single-domain cystatin gives insights on its thermal and pH stability.
Int. J. Biol. Macromol., 102, 2017
|
|
8T3J
 
 | | Crystal structure of native exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Calil, F.A, Gismene, C, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
8T3I
 
 | | Crystal structure of mutant exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Gismene, C, Calil, F.A, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
4U5M
 
 | | Structure of a left-handed DNA G-quadruplex | | Descriptor: | DNA (28-MER), MAGNESIUM ION, POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Brahim, H, Chung, W.J, Lim, K.W. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a left-handed DNA G-quadruplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7D6Z
 
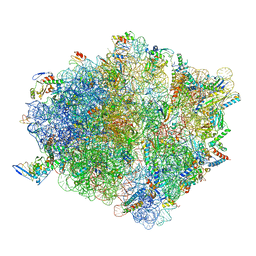 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor | | Descriptor: | 16S ribosomal rRNA, 23S ribosomal rRNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
8TAF
 
 | |
