6HBU
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
6HCO
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to estrone 3-sulfate and 5D3-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
6HS3
 
 | |
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HRC
 
 | | Outward-facing PglK with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, WlaB protein | | Authors: | Perez, C, Locher, K.P. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Outward-Facing PglK and Molecular Dynamics of Lipid-Linked Oligosaccharide Recognition and Translocation.
Structure, 27, 2019
|
|
6JB3
 
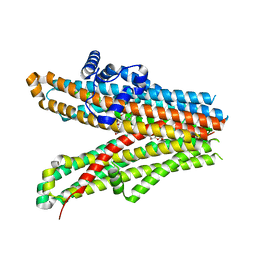 | | Structure of SUR1 subunit bound with repaglinide | | Descriptor: | ATP-binding cassette sub-family C member 8 isoform X2, Digitonin, Repaglinide | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6JBJ
 
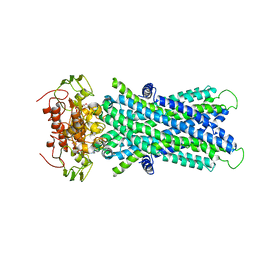 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
6HCF
 
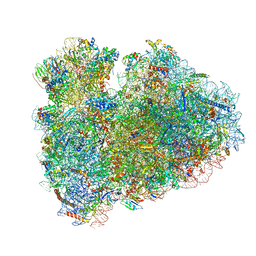 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HZM
 
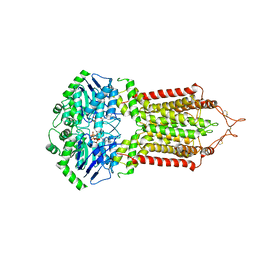 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium (alternative placement of Magnesium into the cryo-EM density) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-10-23 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
6HA8
 
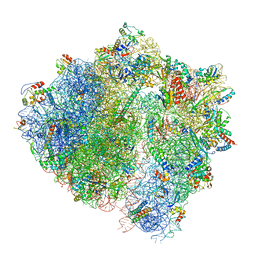 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HIJ
 
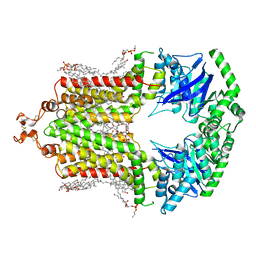 | | Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATP-binding cassette sub-family G member 2, CHOLESTEROL, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-08-30 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat.Struct.Mol.Biol., 25, 2018
|
|
6O1V
 
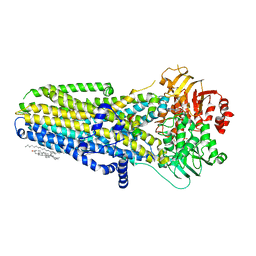 | | Complex of human cystic fibrosis transmembrane conductance regulator (CFTR) and GLPG1837 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J, Levit, A, Shoichet, B. | | Deposit date: | 2019-02-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural identification of a hotspot on CFTR for potentiation.
Science, 364, 2019
|
|
6O30
 
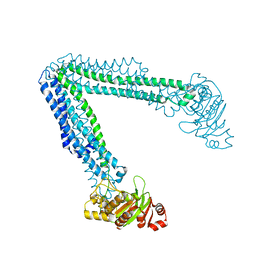 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
6OIH
 
 | |
4M2S
 
 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
4LSG
 
 | |
6PAM
 
 | |
6P6J
 
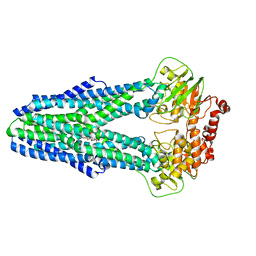 | | Structure of YbtPQ importer with substrate Ybt-Fe bound | | Descriptor: | ABC transporter protein, FE (III) ION, inner membrane ABC-transporter, ... | | Authors: | Wang, Z, Hu, W, Zheng, H. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter.
Sci Adv, 6, 2020
|
|
4M2T
 
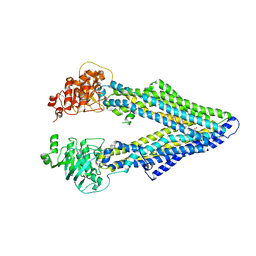 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-SSS | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
6PAN
 
 | |
4MRN
 
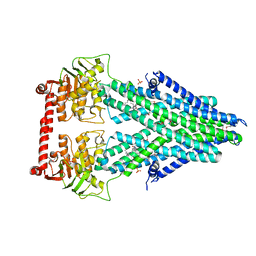 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
6PAQ
 
 | |
6P6I
 
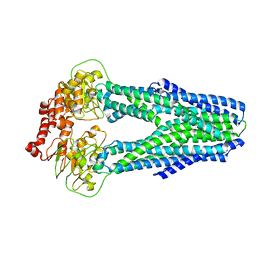 | | Structure of YbtPQ importer | | Descriptor: | ABC transporter protein, inner membrane ABC-transporter | | Authors: | Wang, Z, Hu, W, Zheng, H. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter.
Sci Adv, 6, 2020
|
|
4MRS
 
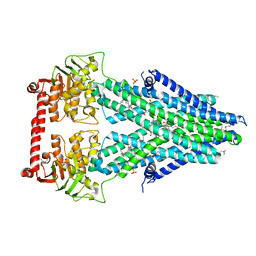 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
6PAO
 
 | |
