4BEK
 
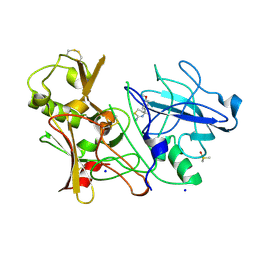 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | (4S)-4-(4-methoxyphenyl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1XOQ
 
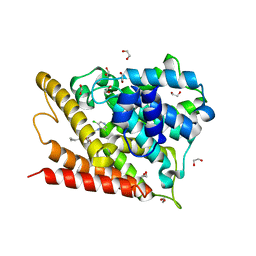 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Roflumilast | | Descriptor: | 1,2-ETHANEDIOL, 3-(CYCLOPROPYLMETHOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-(DIFLUOROMETHOXY)BENZAMIDE, MAGNESIUM ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
7JLK
 
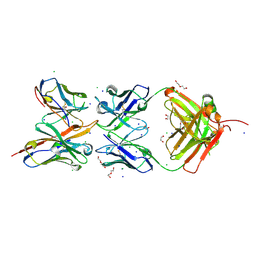 | |
5RHE
 
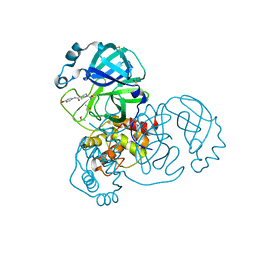 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PG-COV-42 (Mpro-x2052) | | Descriptor: | 1-acetyl-N-(6-methoxypyridin-3-yl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4NZG
 
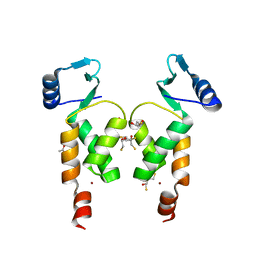 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | Authors: | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
2G95
 
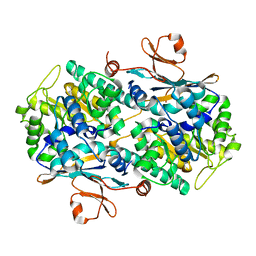 | |
3Q9X
 
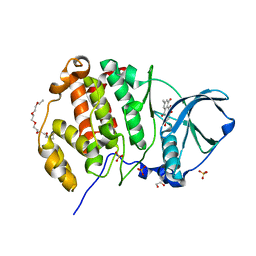 | | Crystal structure of human CK2 alpha in complex with emodin at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, ... | | Authors: | Battistutta, R, Ranchio, A, Papinutto, E. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the flexible regions of the catalytic alpha-subunit of protein kinase CK2
To be Published
|
|
2GCP
 
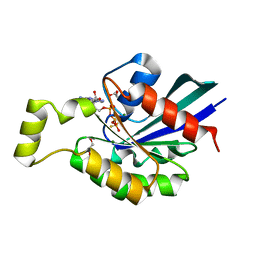 | | Crystal structure of the human RhoC-GSP complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
3RL7
 
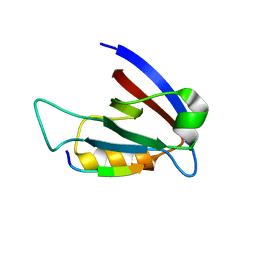 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
2GCH
 
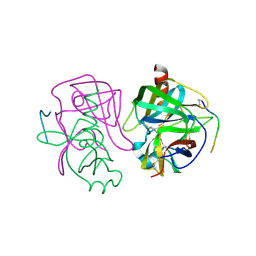 | |
7JOM
 
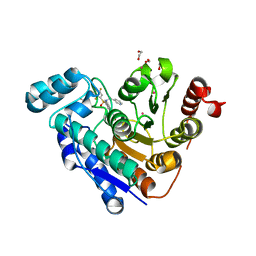 | |
4RDX
 
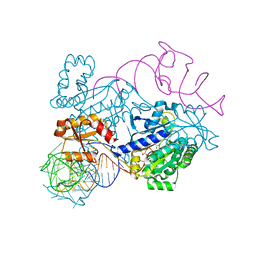 | | Structure of histidinyl-tRNA synthetase in complex with tRNA(His) | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, Histidine--tRNA ligase, ... | | Authors: | Xie, W, Tian, Q, Wang, C. | | Deposit date: | 2014-09-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase
Nucleic Acids Res., 43, 2015
|
|
5EV8
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor D-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4BTL
 
 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
3HBR
 
 | | Crystal structure of OXA-48 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, OXA-48 | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Rossolini, G.M, Docquier, J.D. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases.
Chem.Biol., 16, 2009
|
|
1XPF
 
 | | HIV-1 subtype A genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
4JEY
 
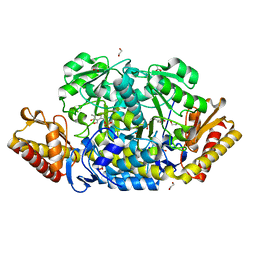 | | E198A mutant of N-acetylornithine aminotransferase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
6MAQ
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside 2'-{[(2S,3R,4R,5R,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-5-nitro-[1,1'-biphenyl]-3-carboxylic acid | | Descriptor: | 2'-{[2-(acetylamino)-2-deoxy-beta-D-galactopyranosyl]oxy}-5-nitro[1,1'-biphenyl]-3-carboxylic acid, Fimbrial adhesin FmlD, GLYCEROL | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
4M1X
 
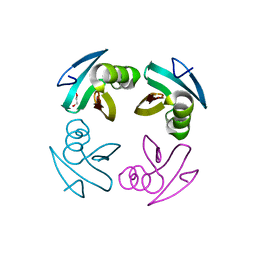 | |
4RJ9
 
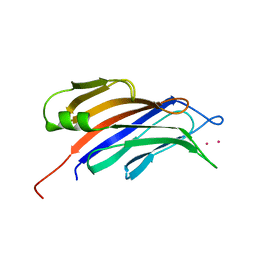 | | Structure of a plant specific C2 domain protein, OsGAP1 from rice | | Descriptor: | C2 domain-containing protein-like, POTASSIUM ION | | Authors: | Miao, R, Fong, Y.H, Wong, K.B, Lam, H.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Site-directed Mutagenesis Shows the Significance of Interactions with Phospholipids and the G-protein OsYchF1 for the Physiological Functions of the Rice GTPase-activating Protein 1 (OsGAP1).
J.Biol.Chem., 290, 2015
|
|
5VSD
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 13 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, GLYCEROL, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
4RJK
 
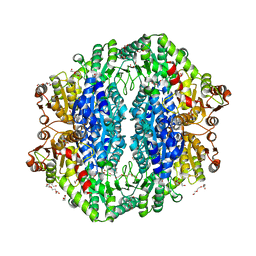 | | Acetolactate synthase from Bacillus subtilis bound to LThDP - crystal form II | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
2GHF
 
 | |
2GDT
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
