2XXM
 
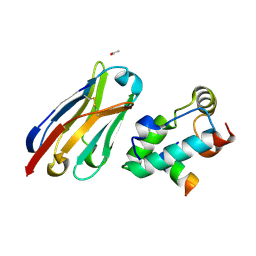 | | Crystal structure of the HIV-1 capsid protein C-terminal domain in complex with a camelid VHH and the CAI peptide. | | Descriptor: | ACETATE ION, CAMELID VHH 9, CAPSID PROTEIN P24, ... | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2W99
 
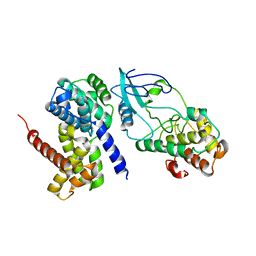 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XMA
 
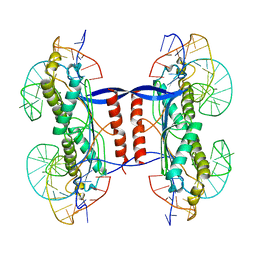 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2NAN
 
 | |
7N2A
 
 | | human PXR LBD bound to compound 2 | | Descriptor: | 5-benzyl-2-(3-fluoro-2-hydroxyphenyl)-6-methyl-3-(2-phenylethyl)pyrimidin-4(3H)-one, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ramanjulu, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTH
 
 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTI
 
 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTB
 
 | |
7N44
 
 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
7OVD
 
 | | Human soluble adenylyl cyclase in complex with the inhibitor TDI10229 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-6-[1,5-dimethyl-4-(phenylmethyl)pyrazol-3-yl]pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Steegborn, C, Quast, J. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of TDI-10229: A Potent and Orally Bioavailable Inhibitor of Soluble Adenylyl Cyclase (sAC, ADCY10).
Acs Med.Chem.Lett., 12, 2021
|
|
7OVY
 
 | | Crystal structure of a dimeric based inhibitor JG34 in complex with the MMP-12 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Ciccone, L, Rossello, A, Vera, L, Nuti, E, Stura, E.A. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7P11
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with quinoline D-galactal ligand | | Descriptor: | 2-[[(2~{R},3~{R},4~{R})-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2~{H}-pyran-4-yl]oxymethyl]quinoline-7-carboxylic acid, CHLORIDE ION, Galectin-8, ... | | Authors: | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | Deposit date: | 2021-07-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
7P1M
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with benzimidazole D-galactal ligand | | Descriptor: | 2-[[(2R,3R,4R)-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2H-pyran-4-yl]oxymethyl]-3-methyl-benzimidazole-5-carboxylic acid, CHLORIDE ION, Galectin-8 | | Authors: | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
7P7P
 
 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
2OJW
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
2OJ0
 
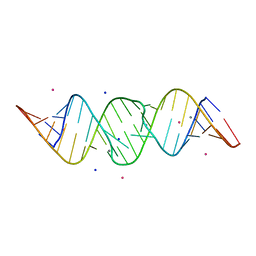 | | Crystal structure of the duplex form of the HIV-1(LAI) RNA dimerization initiation site MN soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2007-01-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cation-dependent cleavage of the duplex form of the subtype-B HIV-1 RNA dimerization initiation site.
Nucleic Acids Res., 38, 2010
|
|
2QC8
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Glutamine synthetase, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-19 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
2PFF
 
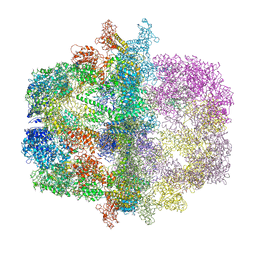 | | Structural Insights of Yeast Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, Tail protein | | Authors: | Xiong, Y, Lomakin, I.B, Steitz, T.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Insights of Yeast Fatty Acid Synthase
Cell(Cambridge,Mass.), 129, 2007
|
|
2OIY
 
 | |
7RUN
 
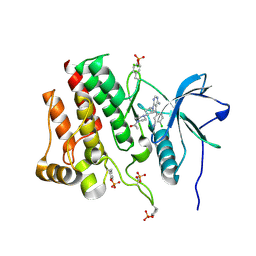 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrrolo[2,3-d]pyrimidine inhibitor. | | Descriptor: | 1-(4-{(1s,3s)-3-[4-amino-5-(3-amino-4-chlorophenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl]cyclobutyl}piperazin-1-yl)ethan-1-one, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2021-08-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Antitarget Selectivity and Tolerability of Novel Pyrrolo[2,3- d ]pyrimidine RET Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
3E26
 
 | |
7SHH
 
 | |
3EQW
 
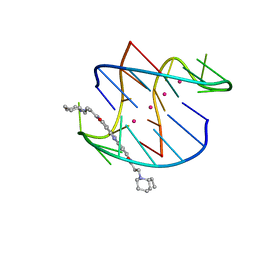 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in small unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3PXO
 
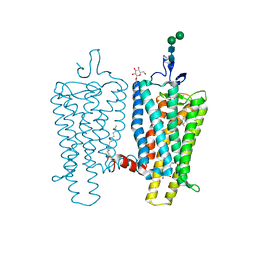 | | Crystal structure of Metarhodopsin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
