3JRW
 
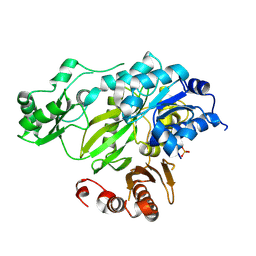 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3K5X
 
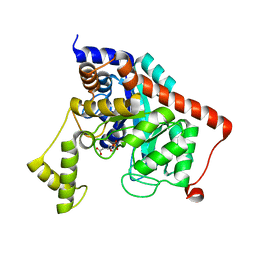 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3CQH
 
 | |
3JSL
 
 | |
3CQR
 
 | |
3K6M
 
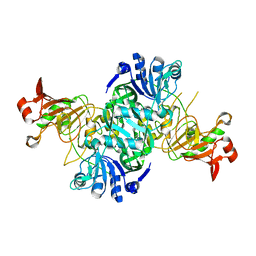 | | Dynamic domains of Succinyl-CoA:3-ketoacid-coenzyme A transferase from pig heart. | | Descriptor: | CHLORIDE ION, GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, ... | | Authors: | Coker, S, Lloyd, A, Mitchell, E, Lewis, G.R, Shoolingin-Jordan, P, Coker, A.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The high-resolution structure of pig heart succinyl-CoA:3-oxoacid coenzyme A transferase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3D0T
 
 | |
3K7G
 
 | | Crystal structure of the Indian Hedgehog N-terminal signalling domain | | Descriptor: | GLYCEROL, Indian hedgehog protein, SULFATE ION, ... | | Authors: | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | Deposit date: | 2009-10-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
3D1E
 
 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | Descriptor: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3JSZ
 
 | | Legionella pneumophila glucosyltransferase Lgt1 N293A with UDP-Glc | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
3CR4
 
 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3JT5
 
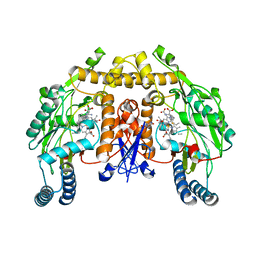 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[2-(ethylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3D23
 
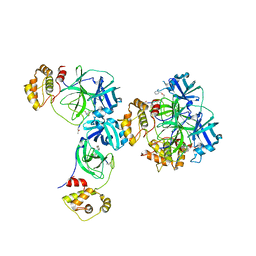 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
3K84
 
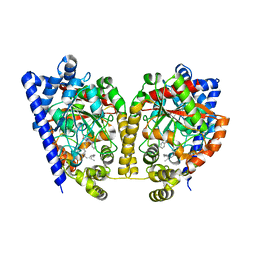 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3D38
 
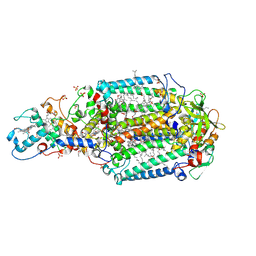 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
3K8T
 
 | | Structure of eukaryotic rnr large subunit R1 complexed with designed adp analog compound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2'-deoxy-2'-(2-hydroxyethyl)adenosine 5'-(trihydrogen diphosphate), GLYCEROL, ... | | Authors: | Sun, D, Xu, H, Dealwis, C, Lee, R.E. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of 2'-(2-Hydroxyethyl)-2'-deoxyadenosine and the 5'-Diphosphate Derivative as Ribonucleotide Reductase Inhibitors
Chemmedchem, 4, 2009
|
|
3JU0
 
 | | Structure of the arm-type binding domain of HAI7 integrase | | Descriptor: | Phage integrase | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
3JU6
 
 | | Crystal Structure of Dimeric Arginine Kinase in Complex with AMPPNP and Arginine | | Descriptor: | ARGININE, Arginine kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, X, Ye, S, Guo, S, Yan, W, Bartlam, M, Rao, Z. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity
Faseb J., 24, 2010
|
|
3CU5
 
 | | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg | | Descriptor: | Two component transcriptional regulator, AraC family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg.
To be Published
|
|
3K9H
 
 | |
3K9O
 
 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
3D52
 
 | | GOLGI MANNOSIDASE II complex with an N-aryl carbamate derivative of gluco-hydroxyiminolactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3JW8
 
 | | Crystal structure of human mono-glyceride lipase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAP
 
 | | Flavodoxin from Desulfovibrio desulfuricans ATCC 27774 (oxidized form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
