8C0F
 
 | | Tubulin-PTC596 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-fluoranyl-2-(6-fluoranyl-2-methyl-benzimidazol-1-yl)-~{N}4-[4-(trifluoromethyl)phenyl]pyrimidine-4,6-diamine, ... | | Authors: | Prota, A.E, Muehlethaler, T, Weetall, M, Steinmetz, M.O. | | Deposit date: | 2022-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1005 Å) | | Cite: | Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Mol Cancer Ther, 20, 2021
|
|
4IO2
 
 | |
1M7G
 
 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M7H
 
 | | Crystal Structure of APS kinase from Penicillium Chrysogenum: Structure with APS soaked out of one dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Adenylylsulfate kinase, ... | | Authors: | Lansdon, E.B, Sege, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
3GCL
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid
J.Biol.Chem., 284, 2009
|
|
4GZF
 
 | | Multi-drug resistant HIV-1 protease 769 variant with reduced LrF peptide | | Descriptor: | LrF peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Kovari, I.A, Brunzelle, J.S, Reiter, S.J, Kovari, L.C. | | Deposit date: | 2012-09-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
5VK4
 
 | |
6K0T
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
1JZM
 
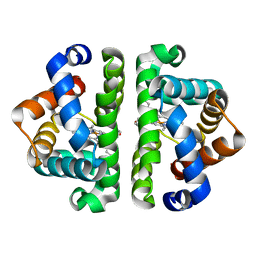 | | Crystal Structure of Scapharca inaequivalvis HbI, I114M Mutant in the Absence of ligand. | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
1FQV
 
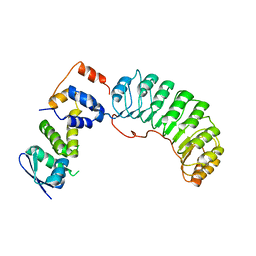 | | Insights into scf ubiquitin ligases from the structure of the skp1-skp2 complex | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-06 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
2M9Y
 
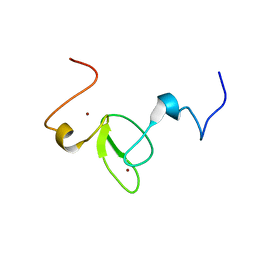 | |
3NM6
 
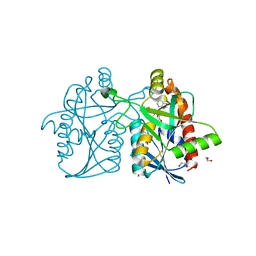 | | Helicobacter pylori MTAN complexed with adenine and tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, ... | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
3NM5
 
 | | Helicobacter pylori MTAN complexed with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
5TJ7
 
 | |
3NM4
 
 | | Helicobacter pylori MTAN | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MTA/SAH nucleosidase | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
3KXW
 
 | |
6T7K
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Wang, J, Tye, M, Payne, N.C, Mazitschek, R, Thompson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
To Be Published
|
|
1YT1
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL | | Descriptor: | Endoplasmin, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
1YSZ
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH NECA | | Descriptor: | Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
3G06
 
 | |
1YT2
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N APO CRYSTAL | | Descriptor: | Endoplasmin, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
2WY3
 
 | | Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Mueller, S, Zocher, G, Steinle, A, Stehle, T. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Hcmv Ul16-Micb Complex Elucidates Select Binding of a Viral Immunoevasin to Diverse Nkg2D Ligands.
Plos Pathog., 6, 2010
|
|
1R6G
 
 | | Crystal structure of the thyroid hormone receptor beta ligand binding domain in complex with a beta selective compound | | Descriptor: | 2-[3,5-DIBROMO-4-(4-HYDROXY-3-{HYDROXY[(2-PHENYLETHYL)AMINO]METHYL}PHENOXY)PHENYL]ETHANE-1,1-DIOL, Thyroid hormone receptor beta-1 | | Authors: | Hangeland, J.J, Dejneka, T, Friends, T.J, Devasthale, P, Mellstrom, K, Sandberg, J, Grynfarb, M, Doweyko, A.M, Sack, J.S, Einspahr, H, Farnegardh, M, Husman, B, Ljunggren, J, Koehler, K, Sheppard, C, Malm, J, Ryono, D.E. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Thyroid receptor ligands. Part 2: Thyromimetics with improved selectivity for the thyroid hormone receptor beta.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3P4E
 
 | | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae.
To be Published
|
|
5SXP
 
 | | STRUCTURAL BASIS FOR THE INTERACTION BETWEEN ITCH PRR AND BETA-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Rho guanine nucleotide exchange factor 7 | | Authors: | Cappadocia, L, Desrochers, G, Lussier-Price, M, Angers, A, Omichinski, J.G. | | Deposit date: | 2016-08-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of interactions between SH3 domain-containing proteins and the proline-rich region of the ubiquitin ligase Itch.
J. Biol. Chem., 292, 2017
|
|
