1L0X
 
 | |
2X8B
 
 | | Crystal structure of human acetylcholinesterase inhibited by aged tabun and complexed with fasciculin-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETHYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Carletti, E, Colletier, J.P, Nachon, F. | | Deposit date: | 2010-03-08 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural evidence that human acetylcholinesterase inhibited by tabun ages through O-dealkylation.
J. Med. Chem., 53, 2010
|
|
182D
 
 | | DNA-NOGALAMYCIN INTERACTIONS: THE CRYSTAL STRUCTURE OF D(TGATCA) COMPLEXED WITH NOGALAMYCIN | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Smith, C.K, Davies, G.J, Dodson, E.J, Moore, M.H. | | Deposit date: | 1994-07-28 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin.
Biochemistry, 34, 1995
|
|
2HQ5
 
 | |
1LEV
 
 | | PORCINE KIDNEY FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH AN AMP-SITE INHIBITOR | | Descriptor: | 3-(2-CARBOXY-ETHYL)-4,6-DICHLORO-1H-INDOLE-2-CARBOXYLIC ACID, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Wright, S.W, Carlo, A.A, Danley, D.E, Hageman, D.L, Karam, G.A, Mansour, M.N, McClure, L.D, Pandit, J, Schulte, G.K, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-04-10 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3-(2-carboxyethyl)-4,6-dichloro-1H-indole-2-carboxylic acid: an allosteric inhibitor of fructose-1,6-bisphosphatase at the AMP site.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2P4P
 
 | | Crystal structure of a CorC_HlyC domain from Haemophilus ducreyi | | Descriptor: | CALCIUM ION, GLYCEROL, Hypothetical protein HD1797, ... | | Authors: | Cuff, M.E, Volkart, L, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-12 | | Release date: | 2007-05-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a CorC_HlyC domain from Haemophilus ducreyi
To be Published
|
|
2AG1
 
 | |
5EXQ
 
 | | Human cytochrome c Y48H | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Fellner, M, Jameson, G.N.L, Ledgerwood, E.C, Wilbanks, S.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity.
Biochem.J., 2021
|
|
6APX
 
 | | Crystal structure of human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the monobody YSX1 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Dual specificity protein phosphatase 1, Monobody YSX1, ... | | Authors: | Gumpena, R, Lountos, G.T, Sreejith, R.K, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Protein Sci., 27, 2018
|
|
2P53
 
 | | Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D273N mutant complexed with N-acetyl phosphonamidate-d-glucosamine-6-phosphate | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-6-O-phosphono-alpha-D-glucopyranose, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-03-14 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Diversity within the Mononuclear and Binuclear Active Sites of N-Acetyl-d-glucosamine-6-phosphate Deacetylase.
Biochemistry, 46, 2007
|
|
1A5A
 
 | | CRYO-CRYSTALLOGRAPHY OF A TRUE SUBSTRATE, INDOLE-3-GLYCEROL PHOSPHATE, BOUND TO A MUTANT (ALPHAD60N) TRYPTOPHAN SYNTHASE ALPHA2BETA2 COMPLEX REVEALS THE CORRECT ORIENTATION OF ACTIVE SITE ALPHA GLU 49 | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE (ALPHA CHAIN), ... | | Authors: | Rhee, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49.
J.Biol.Chem., 273, 1998
|
|
4LXC
 
 | | The antimicrobial peptidase lysostaphin from Staphylococcus simulans | | Descriptor: | Lysostaphin, SULFATE ION, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
3J6T
 
 | | Cryo-EM structure of Dengue virus serotype 3 at 37 degrees C | | Descriptor: | envelope protein, membrane protein | | Authors: | Fibriansah, G, Tan, J.L, Smith, S.A, de Alwis, R, Ng, T.-S, Kostyuchenko, V.A, Kukkaro, P, de Silva, A.M, Crowe Jr, J.E, Lok, S.-M. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins.
Nat Commun, 6, 2015
|
|
1L6M
 
 | | Neutrophil Gelatinase-associated Lipocalin is a Novel Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Goetz, D.H, Borregaard, N, Bluhm, M.E, Raymond, K.N, Strong, R.K. | | Deposit date: | 2002-03-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Neutrophil Lipocalin NGAL is a Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition
Mol.Cell, 10, 2002
|
|
3O60
 
 | |
2WTP
 
 | | Crystal Structure of Cu-form Czce from C. metallidurans CH34 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
2OOI
 
 | |
5F2H
 
 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987
To Be Published
|
|
7CX4
 
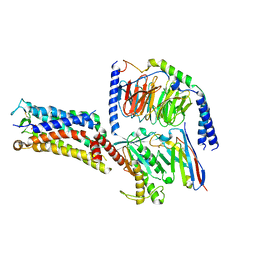 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
1K9W
 
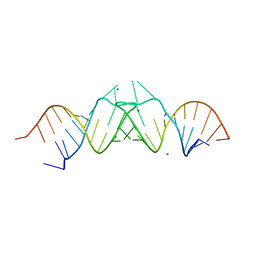 | | HIV-1(MAL) RNA Dimerization Initiation Site | | Descriptor: | HIV-1 DIS(Mal)UU RNA, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site.
Nat.Struct.Biol., 8, 2001
|
|
5BQB
 
 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form III | | Descriptor: | CHLORIDE ION, CITRIC ACID, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
2WYK
 
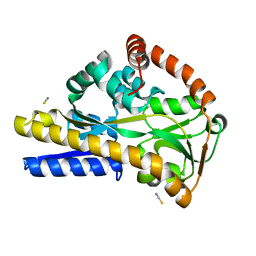 | | SiaP in complex with Neu5Gc | | Descriptor: | N-glycolyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, THIOCYANATE ION | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2009-11-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
2P8P
 
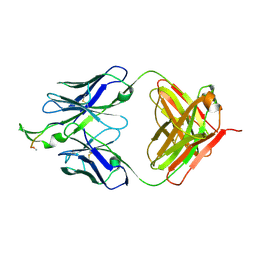 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASLW[N-Ac] | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3EJO
 
 | |
7CX3
 
 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
