5LJ0
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 8-(((3R,4R,5S)-3-((4,4-difluorocyclohexyl)methoxy)-5-methoxypiperidin-4-yl)amino)-3-methyl-5-(5-methylpyridin-3-yl)-1,7-naphthyridin-2(1H)-one | | 分子名称: | 1,2-ETHANEDIOL, 8-(((3R,4R,5S)-3-((4,4-difluorocyclohexyl)methoxy)-5-methoxypiperidin-4-yl)amino)-3-methyl-5-(5-methylpyridin-3-yl)-1,7-naphthyridin-2(1H)-one, ATPase family AAA domain-containing protein 2, ... | | 著者 | Chung, C. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | A Chemical Probe for the ATAD2 Bromodomain.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LKX
 
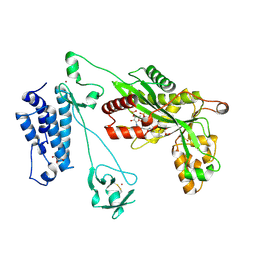 | | Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | 著者 | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | 登録日 | 2016-07-25 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LLY
 
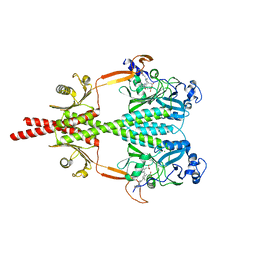 | | Photosensory Module of Bacteriophytochrome linked Diguanylyl Cyclase from Idiomarina species A28L | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, CHLORIDE ION, Diguanylate cyclase (GGDEF) domain-containing protein, ... | | 著者 | Gourinchas, G, Winkler, A. | | 登録日 | 2016-07-28 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Long-range allosteric signaling in red light-regulated diguanylyl cyclases.
Sci Adv, 3, 2017
|
|
5LKU
 
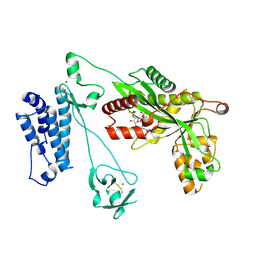 | | Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. | | 分子名称: | COENZYME A, Histone acetyltransferase p300,Histone acetyltransferase p300, ZINC ION | | 著者 | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | 登録日 | 2016-07-25 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LPM
 
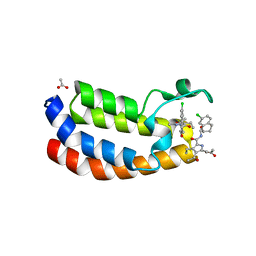 | | Crystal structure of the bromodomain of human Ep300 bound to the inhibitor XDM3d | | 分子名称: | ACETATE ION, Histone acetyltransferase p300, ~{N}-[(1~{S},2~{S})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole -2-carboxamide | | 著者 | Huegle, M, Wohlwend, D, Gerhardt, S. | | 登録日 | 2016-08-13 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7AHO
 
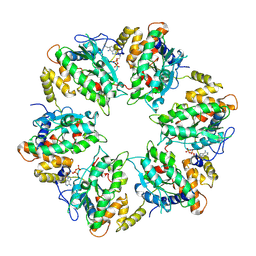 | | RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | 著者 | Lopez-Perrote, A, Rodriguez, C.F, Llorca, O. | | 登録日 | 2020-09-25 | | 公開日 | 2020-11-25 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.
Elife, 9, 2020
|
|
2M0C
 
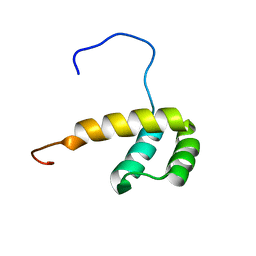 | | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C | | 分子名称: | Homeobox protein aristaless-like 4 | | 著者 | Xu, X, Eletsky, A, Pulavarti, S, Lee, D, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-10-24 | | 公開日 | 2012-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C
To be Published
|
|
7EGF
 
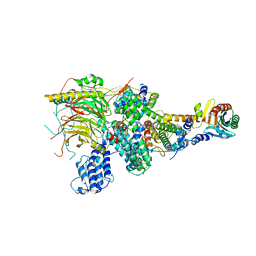 | | TFIID lobe A subcomplex | | 分子名称: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
4YHR
 
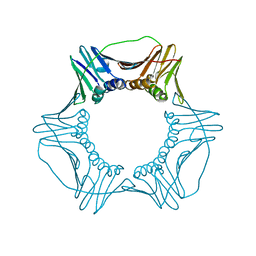 | | Crystal Structure of Yeast Proliferating Cell Nuclear Antigen | | 分子名称: | Proliferating cell nuclear antigen | | 著者 | Litman, J.M, Nguyen, V.Q, Kondratick, C.M, Powers, K.T, Schnieders, M.J, Washington, M.T. | | 登録日 | 2015-02-27 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9502 Å) | | 主引用文献 | Dead-End Elimination with a Polarizable Force Field Repacks PCNA Models from Low-Resolution X-ray Diffraction into Atomic Resolution Structures
To be published
|
|
8PQ4
 
 | |
8CG7
 
 | | Structure of p53 cancer mutant Y220C with arylation at Cys182 and Cys277 | | 分子名称: | 1,2-ETHANEDIOL, 5-iodanyl-2-methylsulfonyl-pyrimidine, Cellular tumor antigen p53, ... | | 著者 | Balourdas, D.I, Pichon, M.M, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-02-03 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structure-Reactivity Studies of 2-Sulfonylpyrimidines Allow Selective Protein Arylation.
Bioconjug.Chem., 34, 2023
|
|
8FFW
 
 | |
8DC6
 
 | |
8DC4
 
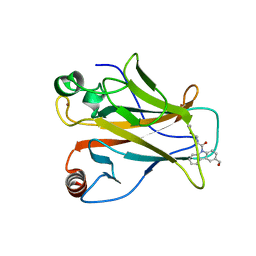 | |
8DC7
 
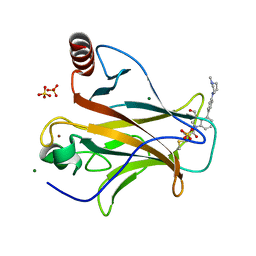 | | Crystal structure of p53 Y220C covalently bound to indole KG10 | | 分子名称: | 4-[4-(4-methylpiperazin-1-yl)phenyl]-1-(2-methylprop-2-enoyl)-1H-indole-3-carbaldehyde, bound form, Cellular tumor antigen p53, ... | | 著者 | Guiley, K.Z, Shokat, K.M. | | 登録日 | 2022-06-15 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9870069 Å) | | 主引用文献 | A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Cancer Discov, 13, 2023
|
|
8DC8
 
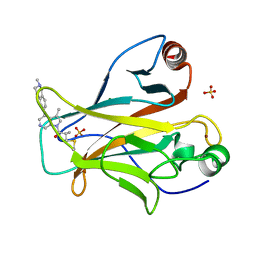 | | Crystal structure of p53 Y220C covalently bound to azaindole KG13 | | 分子名称: | 2-methyl-1-[(4P)-3-methyl-4-(2-methyl-1,2,3,4-tetrahydroisoquinolin-6-yl)-1H-pyrrolo[2,3-c]pyridin-1-yl]prop-2-en-1-one, bound form, Cellular tumor antigen p53, ... | | 著者 | Guiley, K.Z, Shokat, K.M. | | 登録日 | 2022-06-15 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7200973 Å) | | 主引用文献 | A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Cancer Discov, 13, 2023
|
|
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | 分子名称: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | 著者 | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
1JNM
 
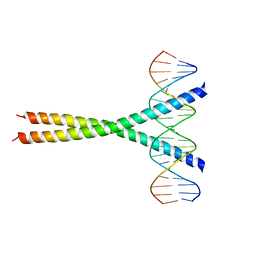 | | Crystal Structure of the Jun/CRE Complex | | 分子名称: | 5'-D(*CP*GP*TP*CP*GP*AP*TP*GP*AP*CP*GP*TP*CP*AP*TP*CP*GP*AP*CP*G)-3', PROTO-ONCOGENE C-JUN | | 著者 | Kim, Y, Podust, L.M. | | 登録日 | 2001-07-24 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Jun bZIP homodimer complexed with CRE
To be Published
|
|
3KOY
 
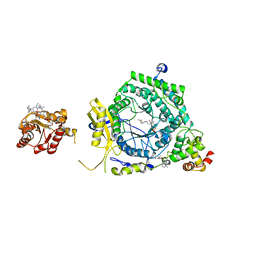 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | 分子名称: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KTA
 
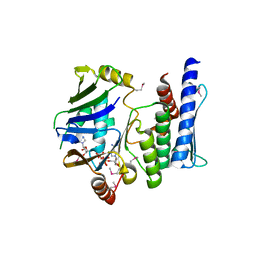 | |
8ZDY
 
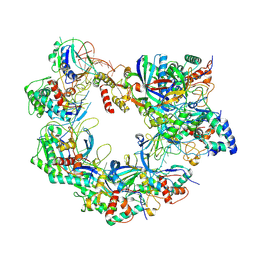 | |
4DMA
 
 | | Crystal structure of ERa LBD in complex with RU100132 | | 分子名称: | 2'-bromo-6'-(furan-3-yl)-4'-(hydroxymethyl)biphenyl-4-ol, Estrogen receptor, Nuclear receptor coactivator 1 | | 著者 | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | 登録日 | 2012-02-07 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ZON
 
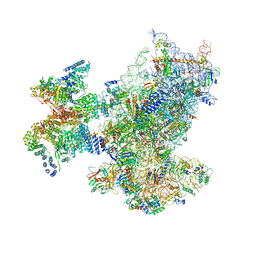 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZN5
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-06 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
