7UMW
 
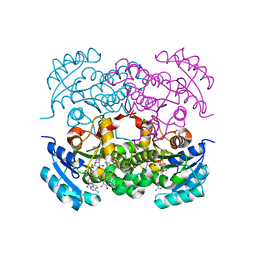 | | Crystal structure of E. Coli FabI in complex with NAD and Fabimycin ((S,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide) | | 分子名称: | (2E)-3-[(7S)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hajian, B. | | 登録日 | 2022-04-08 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
7UTB
 
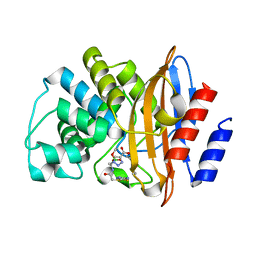 | | KPC-2 CARBAPENEMASE IN COMPLEX WITH THE BORONIC ACID INHIBITOR MB_076 | | 分子名称: | Carbapenem-hydrolyzing beta-lactamase KPC, [(1~{R})-1-[2-[(5-azanyl-1,3,4-thiadiazol-2-yl)sulfanyl]ethanoylamino]-2-(4-carboxy-1,2,3-triazol-1-yl)ethyl]-$l^{3}-oxidanyl-bis(oxidanyl)boron | | 著者 | van den Akker, F, Alsenani, T.A. | | 登録日 | 2022-04-26 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Boronic Acid Transition State Inhibitors as Potent Inactivators of KPC and CTX-M beta-Lactamases: Biochemical and Structural Analyses.
Antimicrob.Agents Chemother., 67, 2023
|
|
5JGU
 
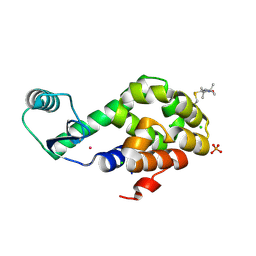 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | 分子名称: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | 著者 | Balo, A.R, Feyrer, H, Ernst, O.P. | | 登録日 | 2016-04-20 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.468 Å) | | 主引用文献 | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
7UJX
 
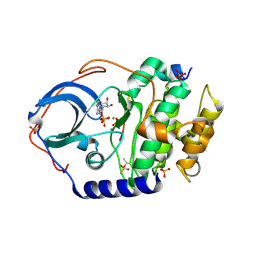 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Wych, D.C, Aoto, P.C, Wall, M.E. | | 登録日 | 2022-03-31 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6WPN
 
 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Substrate-binding protein | | 著者 | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | 登録日 | 2020-04-27 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
5JHA
 
 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin2 | | 分子名称: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)pyrimidin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | 著者 | Burke, J.E, Inglis, A.J, Williams, R.L. | | 登録日 | 2016-04-20 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
6O9E
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA and INDOPY-1 | | 分子名称: | 5-methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ruiz, F.X, Hoang, A, Das, K, Arnold, E. | | 登録日 | 2019-03-13 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1.
J.Med.Chem., 62, 2019
|
|
6O9O
 
 | |
5JHW
 
 | |
6WKO
 
 | |
8JKS
 
 | |
6WXN
 
 | | EGFR(T790M/V948R) in complex with LN3844 | | 分子名称: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | 著者 | Heppner, D.E, Eck, M.J. | | 登録日 | 2020-05-11 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Wych, D.C, Aoto, P.C, Wall, M.E. | | 登録日 | 2022-05-10 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8J7E
 
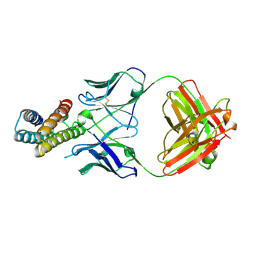 | |
8J3P
 
 | | Formate dehydrogenase mutant from from Candida dubliniensis M4 complexed with NADP+ | | 分子名称: | Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ma, W, Zheng, Y.C, Geng, Q, Chen, C, Xu, J.H. | | 登録日 | 2023-04-17 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
7UYG
 
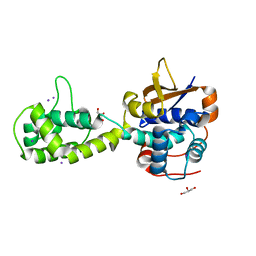 | |
5JJG
 
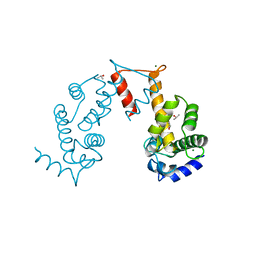 | | Structure of magnesium-loaded ALG-2 | | 分子名称: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Pcalcium-binding protein ALG-2, ... | | 著者 | Tanner, J.J. | | 登録日 | 2016-04-23 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | EF5 Is the High-Affinity Mg(2+) Site in ALG-2.
Biochemistry, 55, 2016
|
|
6OAK
 
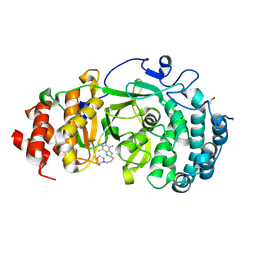 | | Structure of human PARG complexed with JA2131 | | 分子名称: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | 著者 | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | 登録日 | 2019-03-16 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
8J69
 
 | |
6WZP
 
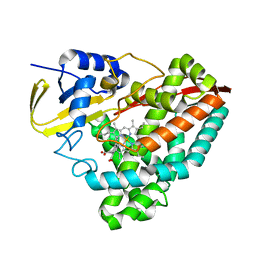 | | The crystal structure of 4-vinylbenzoate-bound T252A mutant CYP199A4 | | 分子名称: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Coleman, T, Bruning, J.B, Bell, S.G. | | 登録日 | 2020-05-14 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
8J1X
 
 | |
7UXO
 
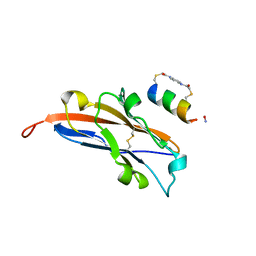 | | Structure of PDL1 in complex with FP30790, a Helicon Polypeptide | | 分子名称: | AMINO GROUP, FP30790, N,N'-(1,4-phenylene)diacetamide, ... | | 著者 | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | 登録日 | 2022-05-05 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UQ3
 
 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and (S)-2-(1-hydroxy-2,5-dioxopyrrolidin-3-yl)acetic acid | | 分子名称: | Bifunctional peptidase and arginyl-hydroxylase JMJD5, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Chowdhury, R, Islam, M.S, Schofield, C.J. | | 登録日 | 2022-04-19 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
8J1W
 
 | |
6X58
 
 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | 分子名称: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | 著者 | McIlwain, B.C, Stockbridge, R.B. | | 登録日 | 2020-05-25 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
