3KSF
 
 | |
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | 著者 | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | 登録日 | 2009-12-06 | | 公開日 | 2010-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
3KLG
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to pre-translocation AZTMP-Terminated DNA (COMPLEX N) | | 分子名称: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), Reverse transcriptase/ribonuclease H, ... | | 著者 | Tu, X, Sarafianos, S.G, Arnold, E. | | 登録日 | 2009-11-07 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | 分子名称: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | 著者 | Tu, X, Sarafianos, S.G, Arnold, E. | | 登録日 | 2009-11-07 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LML
 
 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | 分子名称: | Lin1278 protein | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-01-31 | | 公開日 | 2010-03-09 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
3LNK
 
 | | Structure of BACE bound to SCH743813 | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Orth, P, Cumming, J. | | 登録日 | 2010-02-02 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LP7
 
 | |
3LPL
 
 | | E. coli pyruvate dehydrogenase complex E1 component E571A mutant | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Furey, W. | | 登録日 | 2010-02-05 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQJ
 
 | |
3LQI
 
 | |
3L8V
 
 | |
3LE9
 
 | |
3LCK
 
 | |
3LJD
 
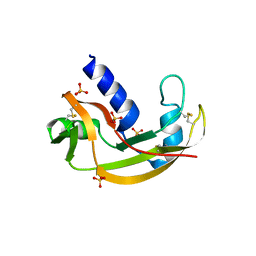 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | 分子名称: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | 著者 | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | 登録日 | 2010-01-26 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LM6
 
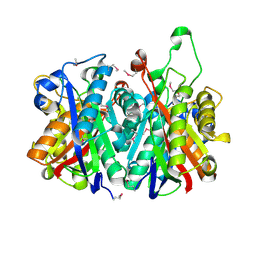 | | Crystal Structure of Stage V sporulation protein AD (spoVAD) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR525 | | 分子名称: | Stage V sporulation protein AD | | 著者 | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-01-29 | | 公開日 | 2010-02-16 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target SR525
To be Published
|
|
3MR3
 
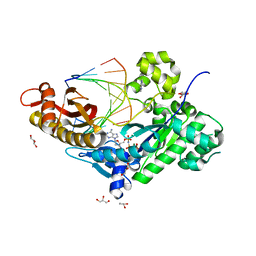 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | 著者 | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | 登録日 | 2010-04-28 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3VX7
 
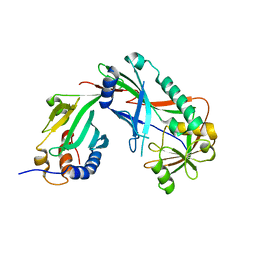 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | 分子名称: | E1, E2 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3W1S
 
 | | Crystal structure of Saccharomyces cerevisiae Atg12-Atg5 conjugate bound to the N-terminal domain of Atg16 | | 分子名称: | Autophagy protein 16, Autophagy protein 5, Ubiquitin-like protein ATG12 | | 著者 | Noda, N.N, Fujioka, Y, Hanada, T, Ohsumi, Y, Inagaki, F. | | 登録日 | 2012-11-20 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Embo Rep., 14, 2013
|
|
3QQK
 
 | | CDK2 in complex with inhibitor L4 | | 分子名称: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](phenyl)methanone | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-02-15 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3MR5
 
 | | Human DNA polymerase eta - DNA ternary complex with a CPD 1bp upstream of the active site (TT3) | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*CP*GP*TP*CP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*(TTD)P*AP*TP*GP*AP*CP*GP*A)-3'), ... | | 著者 | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | 登録日 | 2010-04-28 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3VGO
 
 | |
3VH1
 
 | |
3MDF
 
 | | Crystal structure of the RRM domain of Cyclophilin 33 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase E | | 著者 | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | 登録日 | 2010-03-30 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
3VVM
 
 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | 登録日 | 2012-07-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3VX6
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | 分子名称: | E1 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
