2MC0
 
 | | Structural Basis of a Thiopeptide Antibiotic Multidrug Resistance System from Streptomyces lividans:Nosiheptide in Complex with TipAS | | 分子名称: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, HTH-type transcriptional activator TipA, nosiheptide | | 著者 | Habazettl, J, Allan, M.G, Jensen, P, Sass, H, Grzesiek, S. | | 登録日 | 2013-08-12 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis and dynamics of multidrug recognition in a minimal bacterial multidrug resistance system
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5WK9
 
 | | R186AP450cam with CN and camphor | | 分子名称: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | 著者 | Poulos, T.L, Batabyal, D. | | 登録日 | 2017-07-24 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Effect of Redox Partner Binding on Cytochrome P450 Conformational Dynamics.
J. Am. Chem. Soc., 139, 2017
|
|
5WS2
 
 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, complex with RNA | | 分子名称: | RNA (5'-R(P*AP*AP*AP*AP*A)-3'), Ribonuclease J, SULFATE ION, ... | | 著者 | Li, D.F, Feng, N. | | 登録日 | 2016-12-05 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family
Mol. Microbiol., 106, 2017
|
|
5WTL
 
 | | Crystal structure of the periplasmic portion of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, OmpA family protein, ... | | 著者 | Dai, S, Tan, K, Ye, S, Zhang, R. | | 登録日 | 2016-12-13 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
5ZTA
 
 | | SirB from Bacillus subtilis with Fe3+ | | 分子名称: | FE (III) ION, Sirohydrochlorin ferrochelatase | | 著者 | Fujishiro, T. | | 登録日 | 2018-05-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | A route for metal acquisition for chelatase reaction catalyzed by SirB from Bacillus subtilis
To be published
|
|
6A5Y
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC143 and 9cRA and SRC1 | | 分子名称: | (9cis)-retinoic acid, 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | 著者 | Wang, N, Liu, J. | | 登録日 | 2018-06-25 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
5W7G
 
 | | An envelope of a filamentous hyperthermophilic virus carries lipids in a horseshoe conformation | | 分子名称: | DNA (253-MER), ORF132, ORF140 | | 著者 | Kasson, P, DiMaio, F, Yu, X, Lucas-Staat, S, Krupovic, M, Schouten, S, Prangishvili, D, Egelman, E. | | 登録日 | 2017-06-19 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Model for a novel membrane envelope in a filamentous hyperthermophilic virus.
Elife, 6, 2017
|
|
5WP2
 
 | | 1.44 Angstrom crystal structure of CYP121 from Mycobacterium tuberculosis in complex with substrate and CN | | 分子名称: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, CYANIDE ION, Mycocyclosin synthase, ... | | 著者 | Fielding, A, Dornevil, K, Liu, A. | | 登録日 | 2017-08-03 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.439 Å) | | 主引用文献 | Probing Ligand Exchange in the P450 Enzyme CYP121 from Mycobacterium tuberculosis: Dynamic Equilibrium of the Distal Heme Ligand as a Function of pH and Temperature.
J. Am. Chem. Soc., 139, 2017
|
|
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | 分子名称: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | 著者 | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | 登録日 | 2017-08-03 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.22 Å) | | 主引用文献 | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
2MJ5
 
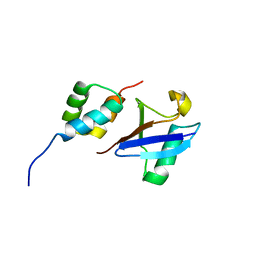 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | 分子名称: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | 著者 | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | 登録日 | 2013-12-25 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
5W9J
 
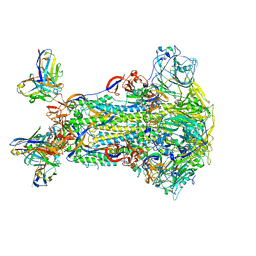 | |
5WTT
 
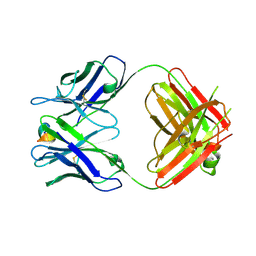 | | Structure of the 093G9 Fab in complex with the epitope peptide | | 分子名称: | Epitope peptide of Cyr61, Heavy chain of 093G9 Fab, Light chain of 093G9 Fab | | 著者 | Zhong, C, Hu, K, Shen, J, Ding, J. | | 登録日 | 2016-12-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-01-02 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for the recognition of CCN1 by monoclonal antibody 093G9.
J. Mol. Recognit., 30, 2017
|
|
6A5X
 
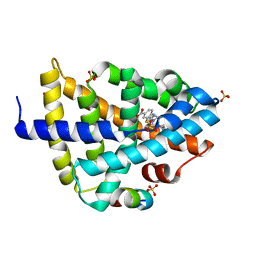 | | FXR-LBD with HNC180 and SRC1 | | 分子名称: | 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | 著者 | Wang, N, Liu, J. | | 登録日 | 2018-06-25 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
5W7K
 
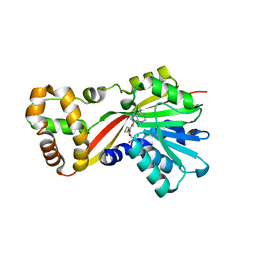 | | Crystal structure of OxaG | | 分子名称: | CHLORIDE ION, OxaG, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W7S
 
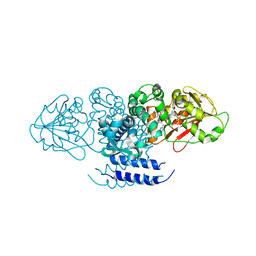 | | Crystal structure of OxaC in complex with sinefungin and meleagrin | | 分子名称: | (3E,7aR,12aS)-6-hydroxy-3-[(1H-imidazol-4-yl)methylidene]-12-methoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, SINEFUNGIN | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.948 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W9P
 
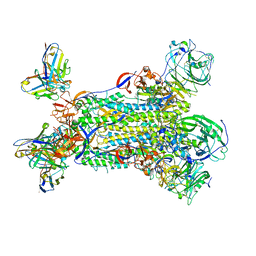 | |
2M8P
 
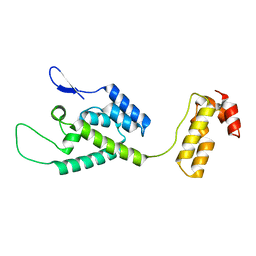 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | 分子名称: | Capsid protein p24 | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | 登録日 | 2013-05-24 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
5WCU
 
 | |
2LXZ
 
 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | 分子名称: | Defensin-5 | | 著者 | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | 登録日 | 2012-09-10 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
5BTP
 
 | |
5W9M
 
 | |
5ZYF
 
 | | Crystal structure of Streptococcus pyogenes type II-A Cas2 | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endoribonuclease Cas2, SULFATE ION | | 著者 | Ka, D, Bae, E. | | 登録日 | 2018-05-24 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2
Nucleic Acids Res., 46, 2018
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | 分子名称: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | 著者 | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | 登録日 | 2012-08-20 | | 公開日 | 2012-09-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
2M8N
 
 | | HIV-1 capsid monomer structure | | 分子名称: | Capsid protein p24 | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | 登録日 | 2013-05-24 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
5WOB
 
 | | Crystal Structure Analysis of Fab1-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Insulin | | 分子名称: | IDE-bound Fab heavy chain, IDE-bound Fab light chain, Insulin, ... | | 著者 | McCord, L.A, Liang, W.G, Farcasanu, M, Wang, A.G, Koide, S, Tang, W.J. | | 登録日 | 2017-08-01 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
