3NCM
 
 | | NEURAL CELL ADHESION MOLECULE, MODULE 2, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (NEURAL CELL ADHESION MOLECULE, LARGE ISOFORM) | | Authors: | Jensen, P.H, Soroka, V, Thomsen, N.K, Berezin, V, Bock, E, Poulsen, F.M. | | Deposit date: | 1998-09-21 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of NCAM modules 1 and 2, basic elements in neural cell adhesion
Nat.Struct.Biol., 6, 1999
|
|
1I2K
 
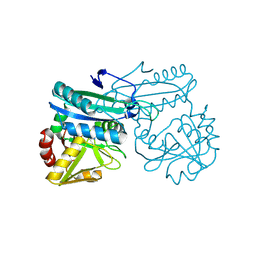 | | AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1I2L
 
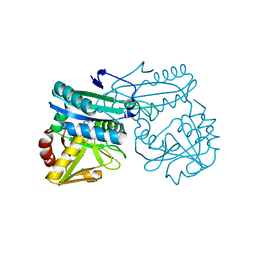 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1ST7
 
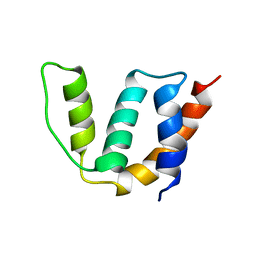 | | Solution structure of Acyl Coenzyme A Binding Protein from yeast | | Descriptor: | Acyl-CoA-binding protein | | Authors: | Teilum, K, Thormann, T, Caterer, N.R, Poulsen, H.I, Jensen, P.H, Knudsen, J, Kragelund, B.B, Poulsen, F.M. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles
Proteins, 59, 2005
|
|
6FHN
 
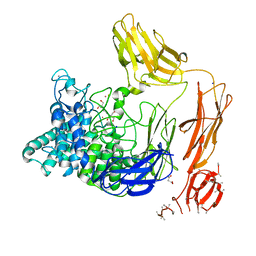 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6FHJ
 
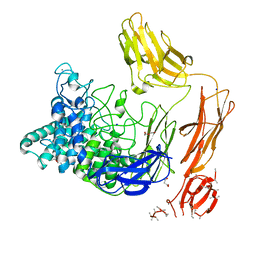 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
2NX6
 
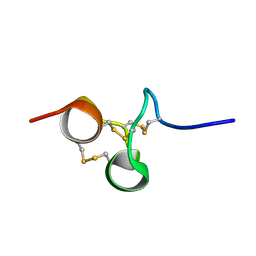 | | Structure of NOWA cysteine rich domain 6 | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Adamczyk, P, Bachinger, H.P, Holstein, T.W, Engel, J, Ozbek, S, Grzesiek, S. | | Deposit date: | 2006-11-17 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequence-structure and structure-function analysis in cysteine-rich domains forming the ultrastable nematocyst wall.
J.Mol.Biol., 368, 2007
|
|
2NX7
 
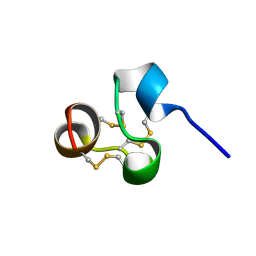 | | Structure of NOWA cysteine rich domain 8 | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Adamczyk, P, Bachinger, H.P, Holstein, T.W, Engel, J, Ozbek, S, Grzesiek, S. | | Deposit date: | 2006-11-17 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequence-structure and structure-function analysis in cysteine-rich domains forming the ultrastable nematocyst wall.
J.Mol.Biol., 368, 2007
|
|
2HM3
 
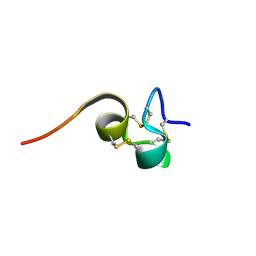 | | Nematocyst outer wall antigen, cysteine rich domain NW1 | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Sequence-Structure and Structure-Function Analysis in Cysteine-rich Domains Forming the Ultrastable Nematocyst Wall.
J.Mol.Biol., 368, 2007
|
|
2MC0
 
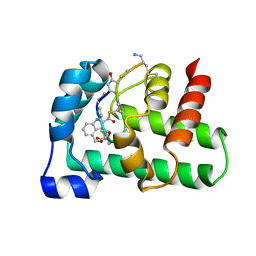 | | Structural Basis of a Thiopeptide Antibiotic Multidrug Resistance System from Streptomyces lividans:Nosiheptide in Complex with TipAS | | Descriptor: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, HTH-type transcriptional activator TipA, nosiheptide | | Authors: | Habazettl, J, Allan, M.G, Jensen, P, Sass, H, Grzesiek, S. | | Deposit date: | 2013-08-12 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and dynamics of multidrug recognition in a minimal bacterial multidrug resistance system
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MBZ
 
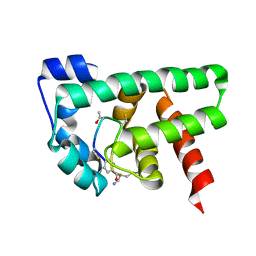 | | Structural Basis of a Thiopeptide Antibiotic Multidrug Resistance System from Streptomyces lividans:Promothiocin A in Complex with TipAS | | Descriptor: | HTH-type transcriptional activator TipA, Promothiocin A | | Authors: | Habazettl, J, Allan, M.G, Jensen, P, Sass, H, Grzesiek, S. | | Deposit date: | 2013-08-12 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and dynamics of multidrug recognition in a minimal bacterial multidrug resistance system.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
1K0G
 
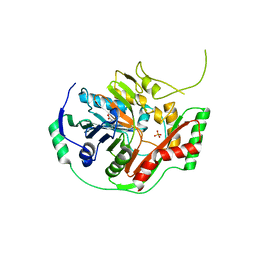 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM PHOSPHATE GROWN CRYSTALS | | Descriptor: | PHOSPHATE ION, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1K0E
 
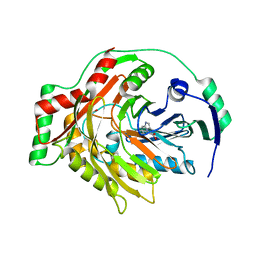 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM FORMATE GROWN CRYSTALS | | Descriptor: | FORMIC ACID, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
2HM4
 
 | | Nematocyst Outer Wall Antigen, NW1 K21P | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Continuous molecular evolution of protein-domain structures by single amino Acid changes.
Curr.Biol., 17, 2007
|
|
2HM6
 
 | | Nematocyst outer wall antigen, NW1 G11V K21P | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Continuous molecular evolution of protein-domain structures by single amino Acid changes.
Curr.Biol., 17, 2007
|
|
2HM5
 
 | | NW1, K21P, Structural Species II | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Continuous molecular evolution of protein-domain structures by single amino Acid changes.
Curr.Biol., 17, 2007
|
|
1LWR
 
 | | Solution structure of the NCAM fibronectin type III module 2 | | Descriptor: | Neural Cell Adhesion Molecule 1, 140 kDa isoform | | Authors: | Kiselyov, V.V, Skladchikova, G, Hinsby, A.M, Jensen, P.H, Kulahin, N, Pedersen, N, Tsetlin, V, Poulsen, F.M, Berezin, V, Bock, E. | | Deposit date: | 2002-06-03 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP
Structure, 11, 2003
|
|
