1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
6SFT
 
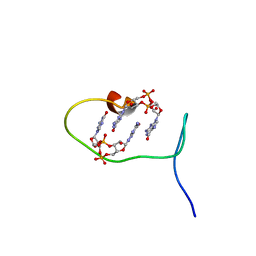 | | Solution structure of protein ARR_CleD in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Two-component receiver protein CleD | | Authors: | Habazettl, J, Hee, C.S, Jenal, U, Schirmer, T, Grzesiek, S. | | Deposit date: | 2019-08-02 | | Release date: | 2020-06-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
2KBL
 
 | |
2L74
 
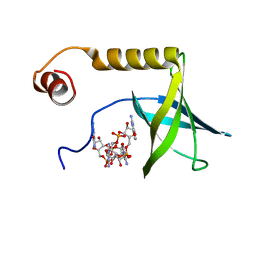 | | Solution structure of the PilZ domain protein PA4608 complex with c-di-GMP identifies charge clustering as molecular readout | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein PA4608 | | Authors: | Habazettl, J, Allan, M, Jenal, U, Grzesiek, S. | | Deposit date: | 2010-12-02 | | Release date: | 2011-02-09 | | Last modified: | 2011-12-28 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout
J.Biol.Chem., 286, 2011
|
|
2MC0
 
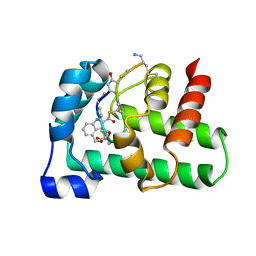 | | Structural Basis of a Thiopeptide Antibiotic Multidrug Resistance System from Streptomyces lividans:Nosiheptide in Complex with TipAS | | Descriptor: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, HTH-type transcriptional activator TipA, nosiheptide | | Authors: | Habazettl, J, Allan, M.G, Jensen, P, Sass, H, Grzesiek, S. | | Deposit date: | 2013-08-12 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and dynamics of multidrug recognition in a minimal bacterial multidrug resistance system
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MBZ
 
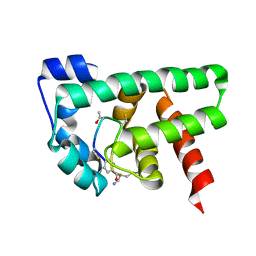 | | Structural Basis of a Thiopeptide Antibiotic Multidrug Resistance System from Streptomyces lividans:Promothiocin A in Complex with TipAS | | Descriptor: | HTH-type transcriptional activator TipA, Promothiocin A | | Authors: | Habazettl, J, Allan, M.G, Jensen, P, Sass, H, Grzesiek, S. | | Deposit date: | 2013-08-12 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and dynamics of multidrug recognition in a minimal bacterial multidrug resistance system.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
3CTI
 
 | |
