8PAV
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | 分子名称: | 1-[3,5-bis(fluoranyl)-4-[[3-(1,3-thiazol-5-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, ... | | 著者 | Friberg, A. | | 登録日 | 2023-06-08 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8JOO
 
 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | 分子名称: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | 著者 | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-08 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8T3W
 
 | | Structure of Bre1-nucleosome complex - state2 | | 分子名称: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | 著者 | Zhao, F, Hicks, C.W, Wolberger, C. | | 登録日 | 2023-06-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3T
 
 | | Structure of Bre1-nucleosome complex - state3 | | 分子名称: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | 著者 | Zhao, F, Hicks, C.W, Wolberger, C. | | 登録日 | 2023-06-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3K
 
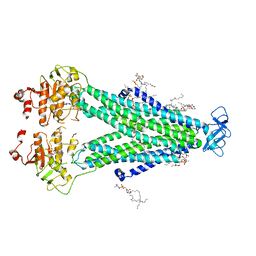 | |
8T3O
 
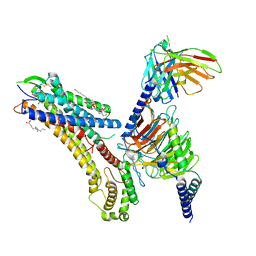 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3Q
 
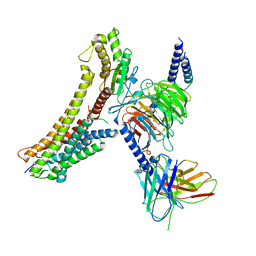 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | 分子名称: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3S
 
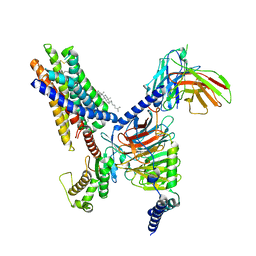 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | 分子名称: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | 分子名称: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3L
 
 | | TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers | | 分子名称: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-06-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8T3M
 
 | | TRPV1 in nanodisc bound with 3 LPA molecules | | 分子名称: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-06-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8JO4
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | 登録日 | 2023-06-07 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Legionella effector LnaB is a phosphoryl-AMPylase that impairs phosphosignalling.
Nature, 2024
|
|
8JO3
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | 登録日 | 2023-06-07 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Legionella effector LnaB is a phosphoryl-AMPylase that impairs phosphosignalling.
Nature, 2024
|
|
8JOH
 
 | |
8T2R
 
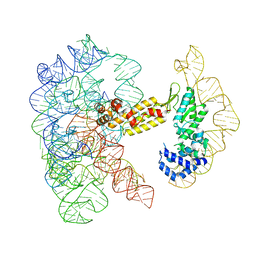 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | 分子名称: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | 著者 | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2T
 
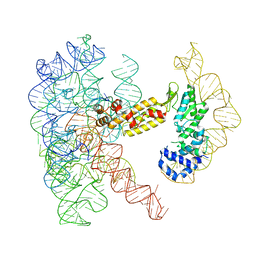 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | 分子名称: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | 著者 | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
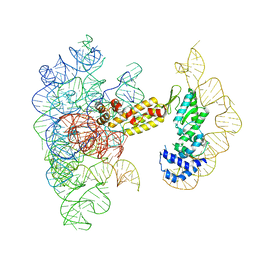 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | 分子名称: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | 著者 | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2H
 
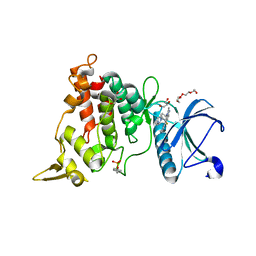 | | DYRK1A complex with DYR530 | | 分子名称: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | 著者 | Montfort, W.R, Basantes, L.E. | | 登録日 | 2023-06-06 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of DYR684, a Potent, Selective, Metabolically Stable, DYRK1A/B PROTAC utilizing a Novel Cereblon Molecular Glue
To Be Published
|
|
8JO2
 
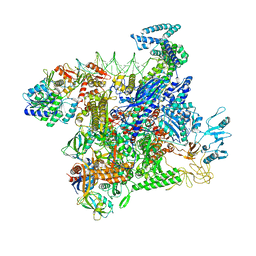 | | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA | | 分子名称: | DNA (65-MER), DNA-binding transcriptional regulator BasR, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lou, Y.-C, Huang, H.-Y, Chen, C, Wu, K.-P. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA.
Nucleic Acids Res., 51, 2023
|
|
8JNC
 
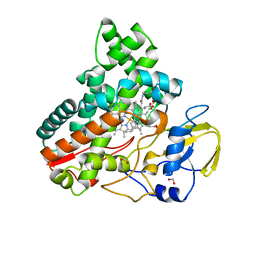 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-maltophilin | | 分子名称: | (1Z,3E,5S,8R,9S,10S,11R,13R,15R,16S,18Z,24S,25S)-11-ethyl-2,24-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-7,20,27,28-tetraone, Cytochrome P450, FORMIC ACID, ... | | 著者 | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNO
 
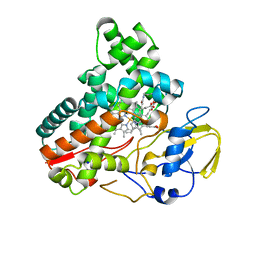 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | 分子名称: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JN8
 
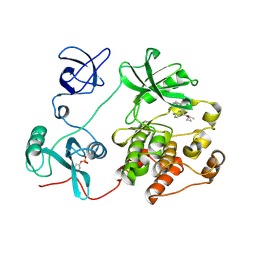 | |
8JN9
 
 | |
8P94
 
 | |
8P9B
 
 | | Crystal Structure of Mnk2-D228G in complex with Tinodasertib | | 分子名称: | 4-[6-(4-morpholin-4-ylcarbonylphenyl)imidazo[1,2-a]pyridin-3-yl]benzenecarbonitrile, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | 著者 | Turnbull, A.P, Sabin, V, Bell, C, Watson, M. | | 登録日 | 2023-06-05 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structure of Mnk2-D228G in complex with Tinodasertib
To Be Published
|
|
