3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | 著者 | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | 登録日 | 2015-10-15 | | 公開日 | 2015-11-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
3JAP
 
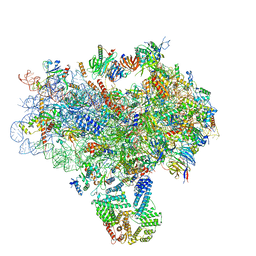 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | 分子名称: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | 著者 | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | 登録日 | 2015-06-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3JAI
 
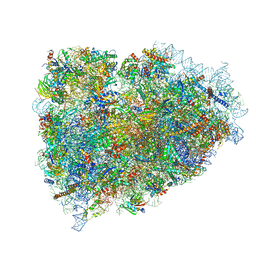 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | 登録日 | 2015-06-10 | | 公開日 | 2015-08-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
3J7R
 
 | | Structure of the translating mammalian ribosome-Sec61 complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | 登録日 | 2014-08-01 | | 公開日 | 2014-09-03 | | 最終更新日 | 2019-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6IP6
 
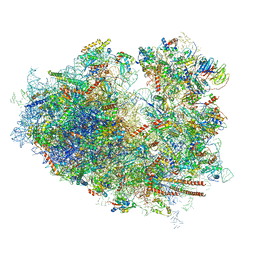 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6KIZ
 
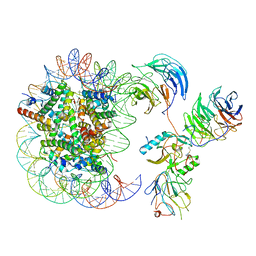 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
3J78
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class I - non-rotated ribosome with 2 tRNAs) | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | 著者 | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2014-05-29 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3J2T
 
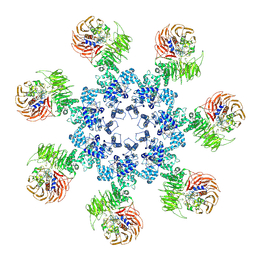 | | An improved model of the human apoptosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | 著者 | Yuan, S, Topf, M, Akey, C.W. | | 登録日 | 2012-12-23 | | 公開日 | 2013-04-10 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Changes in apaf-1 conformation that drive apoptosome assembly.
Biochemistry, 52, 2013
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
3J81
 
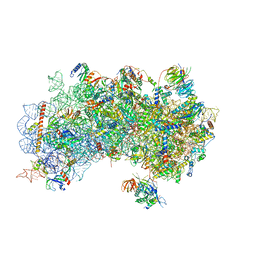 | | CryoEM structure of a partial yeast 48S preinitiation complex | | 分子名称: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | 著者 | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | 登録日 | 2014-08-29 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
3JAH
 
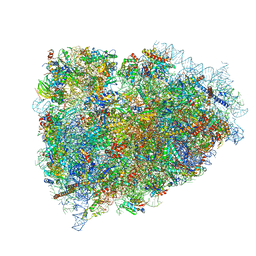 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | 登録日 | 2015-06-10 | | 公開日 | 2015-08-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JCT
 
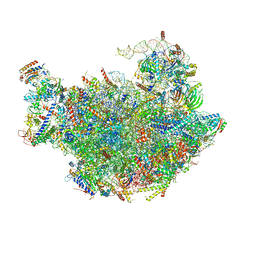 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | 分子名称: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | 登録日 | 2016-03-09 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
3KRX
 
 | |
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KE6
 
 | |
6KIX
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KPF
 
 | | Cryo-EM structure of a class A GPCR with G protein complex | | 分子名称: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | 登録日 | 2019-08-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPG
 
 | | Cryo-EM structure of CB1-G protein complex | | 分子名称: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | 登録日 | 2019-08-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
