5EPY
 
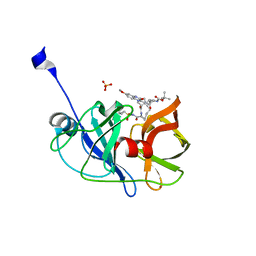 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | 分子名称: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | 著者 | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | 登録日 | 2015-11-12 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EQS
 
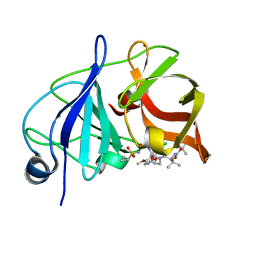 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Asunaprevir | | 分子名称: | N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protease, ZINC ION | | 著者 | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | 登録日 | 2015-11-13 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.839 Å) | | 主引用文献 | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
5EPN
 
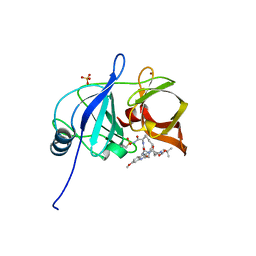 | | Crystal structure of HCV NS3/4A protease in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | 分子名称: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | 著者 | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | 登録日 | 2015-11-11 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
4BJB
 
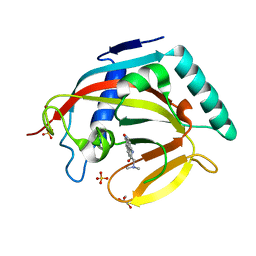 | | Crystal structure of human tankyrase 2 in complex with PJ-34 | | 分子名称: | GLYCEROL, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, SULFATE ION, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2013-04-17 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
5EMS
 
 | | Crystal Structure of an iodinated insulin analog | | 分子名称: | CHLORIDE ION, Insulin, PHENOL, ... | | 著者 | Lawrence, M.C, Pandyarajan, V, Wan, Z, Weiss, M.A. | | 登録日 | 2015-11-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Extending Halogen-based Medicinal Chemistry to Proteins: IODO-INSULIN AS A CASE STUDY.
J. Biol. Chem., 291, 2016
|
|
5WX9
 
 | | Crystal Structure of AtERF96 with GCC-box | | 分子名称: | Ethylene-responsive transcription factor ERF096, GCC-box motif | | 著者 | Chen, C.Y, Cheng, Y.S. | | 登録日 | 2017-01-06 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box.
Plant Mol.Biol., 104, 2020
|
|
7LQR
 
 | | Structure of conotoxin CIC | | 分子名称: | Alpha-conotoxin CIC | | 著者 | Evans, E.R.J, Daly, N.L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-04-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Synthesis, Structural and Pharmacological Characterizations of CIC, a Novel alpha-Conotoxin with an Extended N-Terminal Tail.
Mar Drugs, 19, 2021
|
|
7LQS
 
 | |
5ETX
 
 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-Linear (MK-5172 linear analogue) | | 分子名称: | CHLORIDE ION, NS3 protease, ZINC ION, ... | | 著者 | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | 登録日 | 2015-11-18 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5B3K
 
 | |
1PDZ
 
 | | X-RAY STRUCTURE AND CATALYTIC MECHANISM OF LOBSTER ENOLASE | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, ENOLASE, MANGANESE (II) ION | | 著者 | Janin, J, Duquerroy, S, Camus, C, Le Bras, G. | | 登録日 | 1995-06-05 | | 公開日 | 1995-11-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray structure and catalytic mechanism of lobster enolase.
Biochemistry, 34, 1995
|
|
1PSC
 
 | |
6FZH
 
 | | Crystal structure of a streptococcal dehydrogenase at 1.5 Angstroem resolution | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | 著者 | Gomez, S, Querol-Garcia, J, Sanchez-Barron, G, Subias, M, Gonzalez-Alsina, A, Melchor-Tafur, C, Franco-Hidalgo, V, Alberti, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | 登録日 | 2018-03-14 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Antimicrobials Anacardic Acid and Curcumin Are Not-Competitive Inhibitors of Gram-Positive Bacterial Pathogenic Glyceraldehyde-3-Phosphate Dehydrogenase by a Mechanism Unrelated to Human C5a Anaphylatoxin Binding.
Front Microbiol, 10, 2019
|
|
5B7V
 
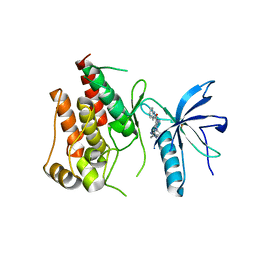 | | Human FGFR1 kinase in complex with CH5183284 | | 分子名称: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | 著者 | Fukami, T.A, Lukacs, C.M, Janson, C. | | 登録日 | 2016-06-09 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
5AWQ
 
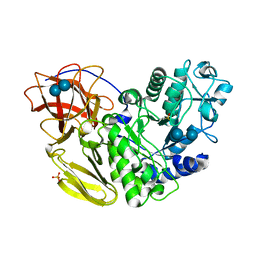 | |
1Q7A
 
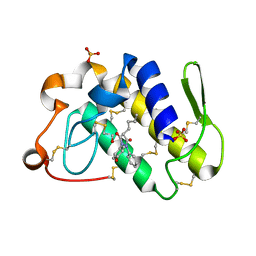 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | 分子名称: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | 著者 | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | 登録日 | 2003-08-17 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
1QAL
 
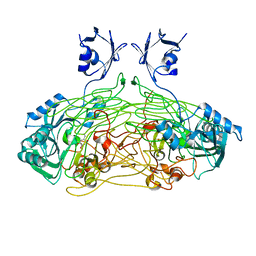 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | 分子名称: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | 著者 | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | 登録日 | 1999-03-19 | | 公開日 | 1999-08-24 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1QBG
 
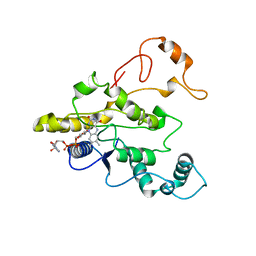 | | CRYSTAL STRUCTURE OF HUMAN DT-DIAPHORASE (NAD(P)H OXIDOREDUCTASE) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | 著者 | Skelly, J.V, Sanderson, M.R, Suter, D.A, Baumann, U, Gregory, D.S, Bennett, M, Hobbs, S.M, Neidle, S. | | 登録日 | 1999-04-20 | | 公開日 | 2000-04-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human DT-diaphorase: a model for interaction with the cytotoxic prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954).
J.Med.Chem., 42, 1999
|
|
7O7Z
 
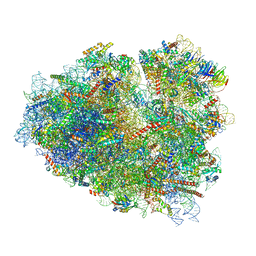 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
5B2X
 
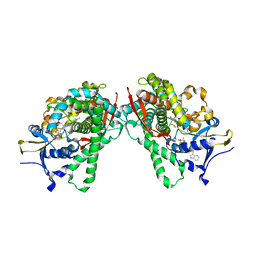 | | Crystal Structure of P450BM3 mutant with N-perfluoroheptanoyl-L-tryptophan | | 分子名称: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,7,7,7-tridecakis(fluoranyl)heptanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | 著者 | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
1QH6
 
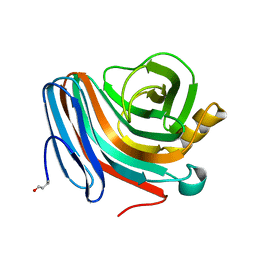 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | 分子名称: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | 著者 | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | 登録日 | 1999-05-11 | | 公開日 | 2000-05-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7AK5
 
 | | Cryo-EM structure of respiratory complex I in the deactive state from Mus musculus at 3.2 A | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | 登録日 | 2020-09-29 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
