6WGM
 
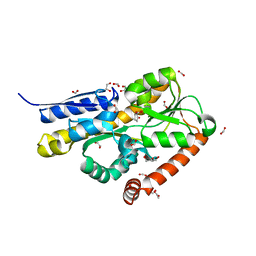 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for pyroglutamate (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, scf7180008839099) in complex with co-purified pyroglutamate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Fedorov, E, Vetting, M.W, Hogle, S.L, Dupont, C.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, GOS_140), in complex with co-purified pyroglutamate
To Be Published
|
|
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | Authors: | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
5IGA
 
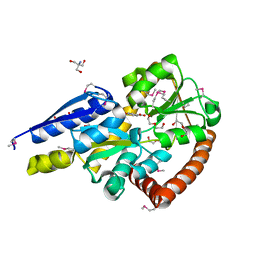 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-27 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate
To be published
|
|
5I7I
 
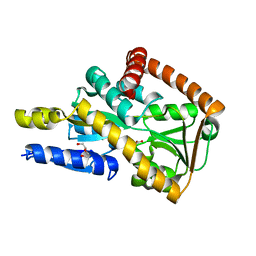 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
5I5P
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
5IZZ
 
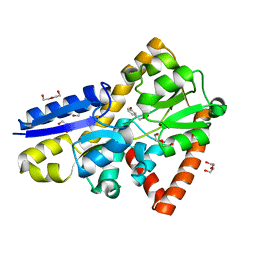 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand | | Descriptor: | 3-HYDROXYPHENYLACETATE, DI(HYDROXYETHYL)ETHER, TRAP TRANSPORTER SOLUTE BINDING PROTEIN | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-03-26 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
To be published
|
|
2C71
 
 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a magnesium ion. | | Descriptor: | GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-17 | | Release date: | 2006-01-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell-Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2C79
 
 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a colbalt ion. | | Descriptor: | COBALT (II) ION, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2CC0
 
 | | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate | | Descriptor: | ACETATE ION, ACETYL-XYLAN ESTERASE, ZINC ION | | Authors: | Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Vincent, F, Brzozowski, A.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Puchart, V, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases
J.Biol.Chem., 281, 2006
|
|
2NLR
 
 | |
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1CWW
 
 | | SOLUTION STRUCTURE OF THE CASPASE RECRUITMENT DOMAIN (CARD) FROM APAF-1 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Day, C.L, Dupont, C, Lackmann, M, Vaux, D.L, Hinds, M.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-01-21 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1.
Cell Death Differ., 6, 1999
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1QH7
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1QH6
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
