5Y28
 
 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | 分子名称: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | 著者 | Zhang, Z, Huang, Q, Tao, X. | | 登録日 | 2017-07-24 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.08606815 Å) | | 主引用文献 | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
5Y3J
 
 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and TCGCAC DNA) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Ohto, U, Ishida, H, Shimizu, T. | | 登録日 | 2017-07-29 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
2QD1
 
 | | 2.2 Angstrom Structure of the human ferrochelatase variant E343K with substrate bound | | 分子名称: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | 著者 | Medlock, A.E, Dailey, T.A, Ross, T.A, Dailey, H.A, Lanzilotta, W.N. | | 登録日 | 2007-06-20 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A pi-Helix Switch Selective for Porphyrin Deprotonation and Product Release in Human Ferrochelatase.
J.Mol.Biol., 373, 2007
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | 分子名称: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | 著者 | Zeqiraj, E, Judd, A, Sicheri, F. | | 登録日 | 2014-06-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7FJP
 
 | | Cryo EM structure of lysosomal ATPase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Zhang, S.S. | | 登録日 | 2021-08-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7FJQ
 
 | | Cryo EM structure of lysosomal ATPase | | 分子名称: | Polyamine-transporting ATPase 13A2, SPERMINE | | 著者 | Zhang, S.S. | | 登録日 | 2021-08-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7YEH
 
 | | Cryo-EM structure of human OGT-OGA complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | 著者 | Lu, P, Liu, Y, Yu, H, Gao, H. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
7YEA
 
 | | Human O-GlcNAc transferase Dimer | | 分子名称: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Gao, H, Lu, P, Liu, Y. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
8JBM
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mn2+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBL
 
 | | Crystal structure of Na+,K+-ATPase in the E1.Mg2+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBK
 
 | | Crystal structure of Na+,K+-ATPase in the E1.3Na+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
6RIP
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in swiveled state | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-24 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RI9
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in non-swiveled state | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-23 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | 分子名称: | CHLORIDE ION, Catalase, GLYCEROL, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.895 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
7M8V
 
 | | Human CYP11B2 in complex with LCI699 | | 分子名称: | 4-[(4R,5R)-6,7-dihydro-5H-pyrrolo[1,2-c]imidazol-5-yl]-3-fluorobenzonitrile, Cytochrome P450 11B2, mitochondrial, ... | | 著者 | Scott, E.E, Brixius-Anderko, S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Aldosterone Synthase Structure With Cushing Disease Drug LCI699 Highlights Avenues for Selective CYP11B Drug Design.
Hypertension, 78, 2021
|
|
6JBQ
 
 | |
6P7X
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6JNX
 
 | | Cryo-EM structure of a Q-engaged arrested complex | | 分子名称: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Feng, Y, Shi, J. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.08 Å) | | 主引用文献 | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
5YTO
 
 | |
6RIN
 
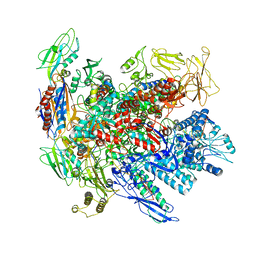 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-24 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6C5W
 
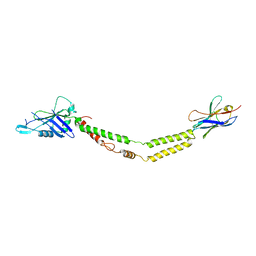 | | Crystal structure of the mitochondrial calcium uniporter | | 分子名称: | CALCIUM ION, calcium uniporter, nanobody | | 著者 | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | 登録日 | 2018-01-17 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.10010242 Å) | | 主引用文献 | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
5YUL
 
 | |
6DDA
 
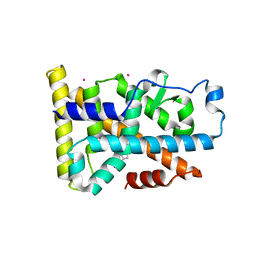 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | 分子名称: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | 著者 | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | 登録日 | 2018-05-09 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
8CE0
 
 | | N-terminal domain of human apolipoprotein E | | 分子名称: | GLYCEROL, Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
8CDY
 
 | | N-terminal domain of human apolipoprotein E | | 分子名称: | Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
