5CTN
 
 | | Structure of BPu1 beta-lactamase | | 分子名称: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, CITRATE ANION | | 著者 | Smith, C.A, Vakulenko, S.B. | | 登録日 | 2015-07-24 | | 公開日 | 2015-11-25 | | 最終更新日 | 2016-10-05 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5DVP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with Doripenem adduct | | 分子名称: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, L,D-transpeptidase 2, PHOSPHONOACETALDEHYDE, ... | | 著者 | Kumar, P, Lamichhane, G. | | 登録日 | 2015-09-21 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5YL3
 
 | | Crystal structure of horse heart myoglobin reconstituted with manganese porphycene in resting state at pH 8.5 | | 分子名称: | Myoglobin, PORPHYCENE CONTAINING MN, SULFATE ION | | 著者 | Oohora, K, Meichin, H, Kihira, Y, Sugimoto, H, Shiro, Y, Hayashi, T. | | 登録日 | 2017-10-17 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Manganese(V) Porphycene Complex Responsible for Inert C-H Bond Hydroxylation in a Myoglobin Matrix.
J. Am. Chem. Soc., 139, 2017
|
|
6PXX
 
 | | Class D beta-lactamase in complex with beta-lactam antibiotic | | 分子名称: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | van den Akker, F, Kumar, V. | | 登録日 | 2019-07-28 | | 公開日 | 2019-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Analysis of The OXA-48 Carbapenemase Bound to A "Poor" Carbapenem Substrate, Doripenem.
Antibiotics, 8, 2019
|
|
3IQA
 
 | | Crystal Structure of BlaC covalently bound with Doripenem | | 分子名称: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Tremblay, L.W, Blanchard, J.S. | | 登録日 | 2009-08-19 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
3ISG
 
 | | Structure of the class D beta-lactamase OXA-1 in complex with doripenem | | 分子名称: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-1 | | 著者 | Powers, R.A. | | 登録日 | 2009-08-25 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The 1.4 A crystal structure of the class D beta-lactamase OXA-1 complexed with doripenem.
Biochemistry, 48, 2009
|
|
6PW8
 
 | |
3WJM
 
 | | Crystal structure of Bombyx mori Sp2/Sp3 heterohexamer | | 分子名称: | Arylphorin, Silkworm storage protein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yuan, Y.A, Hou, Y. | | 登録日 | 2013-10-11 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Bombyx mori arylphorins reveals a 3:3 heterohexamer with multiple papain cleavage sites
Protein Sci., 23, 2014
|
|
5JFT
 
 | | Zebra Fish Caspase-3 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACE-ASP-GLU-VAL-ASK, ... | | 著者 | Tucker, M.B, MacKenzie, S.H, Maciag, J.J, Dirscherl, H, Swartz, P.D, Yoder, J.A, Hamilton, P.T, Clark, A.C. | | 登録日 | 2016-04-19 | | 公開日 | 2016-10-26 | | 最終更新日 | 2016-11-02 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish.
Protein Sci., 25, 2016
|
|
1N4K
 
 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | 登録日 | 2002-10-31 | | 公開日 | 2002-12-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
2K87
 
 | | NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS | | 分子名称: | Non-structural protein 3 of Replicase polyprotein 1a | | 著者 | Serrano, P, Wuthrich, K, Johnson, M.A, Chatterjee, A, Wilson, I, Pedrini, B.F, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-09-02 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3.
J.Virol., 83, 2009
|
|
3T8S
 
 | | Apo and InsP3-bound Crystal Structures of the Ligand-Binding Domain of an InsP3 Receptor | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Lin, C, Baek, K, Lu, Z. | | 登録日 | 2011-08-01 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.77 Å) | | 主引用文献 | Apo and InsP(3)-bound crystal structures of the ligand-binding domain of an InsP(3) receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2WCT
 
 | | human SARS coronavirus unique domain (triclinic form) | | 分子名称: | NON-STRUCTURAL PROTEIN 3 | | 著者 | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2009-03-16 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
4GUA
 
 | | Alphavirus P23pro-zbd | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural polyprotein, SULFATE ION, ... | | 著者 | Shin, G, Yost, S, Miller, M, Marcotrigiano, J. | | 登録日 | 2012-08-29 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.854 Å) | | 主引用文献 | Structural and functional insights into alphavirus polyprotein processing and pathogenesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | 分子名称: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | 著者 | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | 登録日 | 2024-03-11 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.395 Å) | | 主引用文献 | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | 分子名称: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | 著者 | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | 登録日 | 2022-04-28 | | 公開日 | 2022-12-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | 著者 | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | 登録日 | 2022-04-28 | | 公開日 | 2022-12-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | 著者 | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | 登録日 | 2022-04-28 | | 公開日 | 2022-12-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
5NRL
 
 | | Structure of a pre-catalytic spliceosome. | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | 著者 | Plaschka, C, Lin, P.-C, Nagai, K. | | 登録日 | 2017-04-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Structure of a pre-catalytic spliceosome.
Nature, 546, 2017
|
|
8CB3
 
 | | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Papain-like protease nsp3 | | 著者 | Morin, B, Ferron, F, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | 登録日 | 2023-01-25 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.572 Å) | | 主引用文献 | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid
To Be Published
|
|
5UTV
 
 | |
8CX9
 
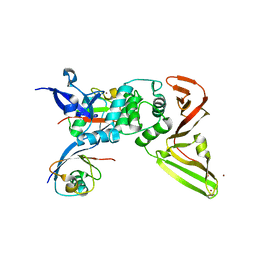 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | 分子名称: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | 著者 | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | 登録日 | 2022-05-20 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
8FWN
 
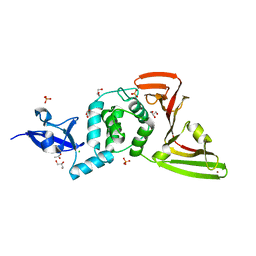 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | 登録日 | 2023-01-23 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
8HDA
 
 | |
2N1U
 
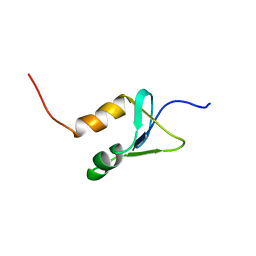 | | Structure of SAP30L corepressor protein | | 分子名称: | Histone deacetylase complex subunit SAP30L, ZINC ION | | 著者 | Tossavainen, H, Permi, P. | | 登録日 | 2015-04-23 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function.
Protein Sci., 25, 2016
|
|
