3LJZ
 
 | | Crystal Structure of Human MMP-13 complexed with an Amino-2-indanol compound | | 分子名称: | (2R)-2-[4-(1,3-benzodioxol-5-yl)benzyl]-N~4~-hydroxy-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Shieh, H.-S, Kiefer, J.R. | | 登録日 | 2010-01-26 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure analysis reveals the flexibility of the ADAMTS-5 active site.
Protein Sci., 20, 2011
|
|
6DTW
 
 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | 分子名称: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | 登録日 | 2018-06-18 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.742 Å) | | 主引用文献 | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5DLS
 
 | | Identification of Novel, in vivo Active Chk1 Inhibitors Utilizing Structure Guided Drug Design | | 分子名称: | 1-benzyl-N-(5-{5-[3-(dimethylamino)-2,2-dimethylpropoxy]-1H-indol-2-yl}-6-oxo-1,6-dihydropyridin-3-yl)-1H-pyrazole-4-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | 著者 | Massey, A.J, Stokes, S, Browne, H, Foloppe, N, Fiumana, A, Scrace, S, Fallowfield, M, Bedford, S, Webb, P, Baker, L.M, Christie, M, Drysdale, M.J, Wood, M. | | 登録日 | 2015-09-07 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification of novel, in vivo active Chk1 inhibitors utilizing structure guided drug design.
Oncotarget, 6, 2015
|
|
6DH1
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with UMass1 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.971 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
5BUK
 
 | | Structure of flavin-dependent chlorinase Mpy16 | | 分子名称: | FADH2-dependent halogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | 登録日 | 2015-06-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6B16
 
 | | P21-activated kinase 1 in complex with a 4-azaindole inhibitor | | 分子名称: | N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-[(1S)-1-(1H-pyrrolo[3,2-b]pyridin-5-yl)ethyl]pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PAK 1 | | 著者 | Rouge, L, Wang, W. | | 登録日 | 2017-09-16 | | 公開日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.285 Å) | | 主引用文献 | Synthesis and evaluation of a series of 4-azaindole-containing p21-activated kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | 分子名称: | Temporin-B | | 著者 | Manzo, G, Mason, J.A. | | 登録日 | 2018-05-12 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | 分子名称: | Ubiquitin D | | 著者 | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | 登録日 | 2018-04-29 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GIJ
 
 | |
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-12 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3HZG
 
 | | Crystal structure of mycobacterium tuberculosis thymidylate synthase X bound with FAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Staker, B.L, Rathod, P, Hunter, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2009-06-23 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
5BSA
 
 | |
7Y3J
 
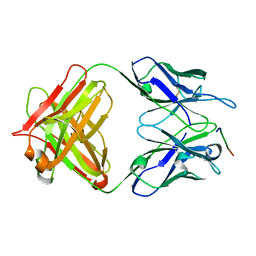 | | 24B3 antibody-peptide complex | | 分子名称: | 24B3 Heavy chain, 24B3 Light chain, ALA-LEU-VAL-PHE-PHE-ALA-PRO-ALA-VAL-GLY-SER | | 著者 | Irie, K, Irie, Y, Kita, A, Miki, K. | | 登録日 | 2022-06-11 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of the 24B3 antibody against the toxic conformer of amyloid beta with a turn at positions 22 and 23.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
3HWI
 
 | |
7YKN
 
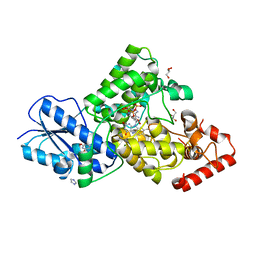 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | 登録日 | 2022-07-23 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | 分子名称: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2010-01-09 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
6DGX
 
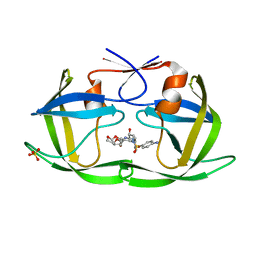 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH6
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | 著者 | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | 登録日 | 2018-06-13 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
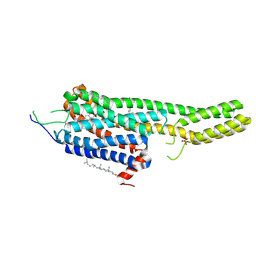 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | 著者 | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | 登録日 | 2018-06-13 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.918 Å) | | 主引用文献 | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7Y43
 
 | |
5EAE
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor R-desthio-prothioconazole | | 分子名称: | (2~{R})-2-(1-chloranylcyclopropyl)-1-(2-chlorophenyl)-3-(1,2,4-triazol-1-yl)propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | 登録日 | 2015-10-16 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
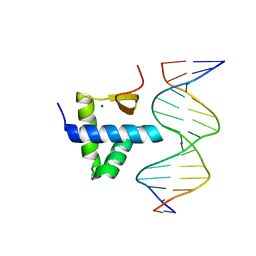
7XI3
 
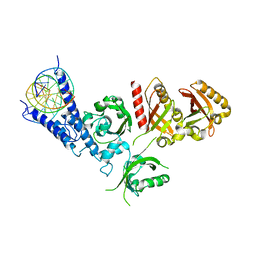 | | Crystal Structure of the NPAS4-ARNT2 heterodimer in complex with DNA | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator 2, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | 著者 | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | 登録日 | 2022-04-11 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (4.274 Å) | | 主引用文献 | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DGY
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass1 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH7
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass1 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
