6BN7
 
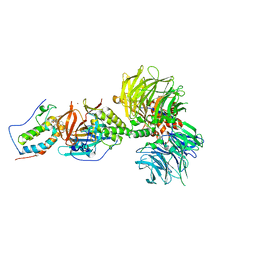 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BOY
 
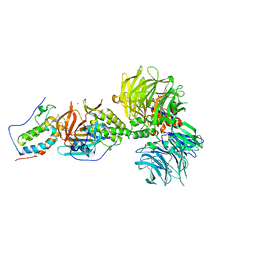 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | 分子名称: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-21 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (6.343 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6WCQ
 
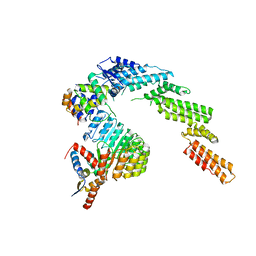 | | Structure of a substrate-bound DQC ubiquitin ligase | | 分子名称: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | 著者 | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | 登録日 | 2020-03-31 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6BN8
 
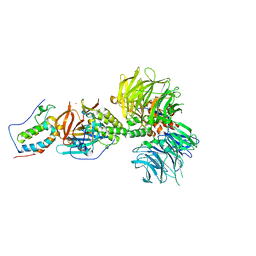 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.990035 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
5IG9
 
 | |
5T5S
 
 | | A fragment of a human tRNA synthetase | | 分子名称: | Alanine--tRNA ligase, cytoplasmic | | 著者 | Sun, L, Schimmel, P. | | 登録日 | 2016-08-31 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8AJN
 
 | | Structure of the human DDB1-DCAF12 complex | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJM
 
 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (30.6 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | 分子名称: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | 分子名称: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | 著者 | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-04-08 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | 登録日 | 2023-12-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
3ASK
 
 | | Structure of UHRF1 in complex with histone tail | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | 著者 | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | 登録日 | 2010-12-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2013-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | 分子名称: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
7K79
 
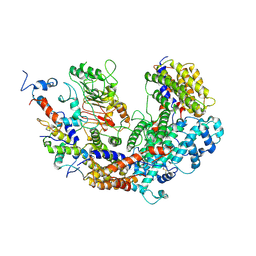 | | CBF3 | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | 著者 | Ruifang, G, Yawen, B. | | 登録日 | 2020-09-22 | | 公開日 | 2021-03-31 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | 分子名称: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | 著者 | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-02-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (4.802 Å) | | 主引用文献 | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
4P5O
 
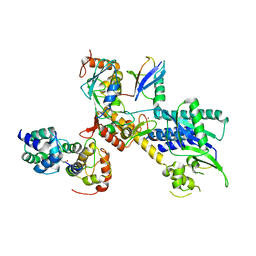 | |
5AIT
 
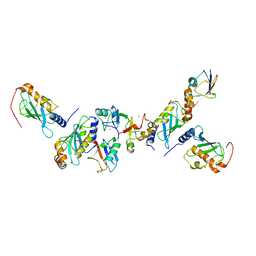 | | A complex of of RNF4-RING domain, UbeV2, Ubc13-Ub (isopeptide crosslink) | | 分子名称: | E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 N, ... | | 著者 | Branigan, E, Naismith, J.H. | | 登録日 | 2015-02-17 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
6GYU
 
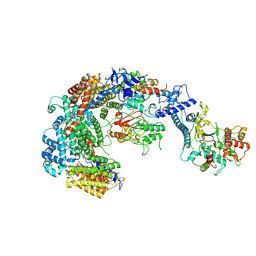 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
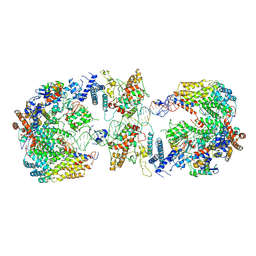 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
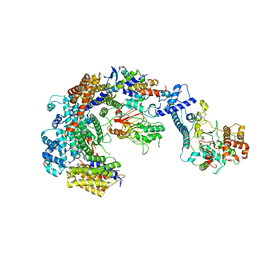 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | 分子名称: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7ZN7
 
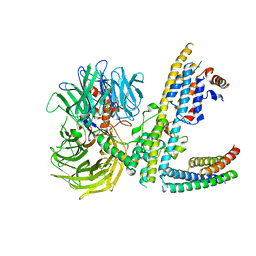 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-20 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
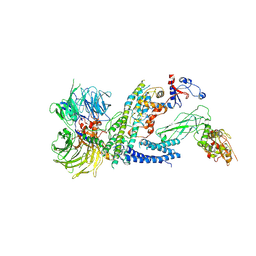 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
6F07
 
 | | CBF3 Core Complex | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit B, Suppressor of kinetochore protein 1, ZINC ION | | 著者 | Leber, V, Singleton, M.R. | | 登録日 | 2017-11-17 | | 公開日 | 2017-12-13 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for assembly of the CBF3 kinetochore complex.
EMBO J., 37, 2018
|
|
