7MEB
 
 | | CDD-1 beta-lactamase in imidazole/MPD 2 minute avibactam complex | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | 著者 | Smith, C.A, Vakulenko, S.B. | | 登録日 | 2021-04-06 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEH
 
 | | CDD-1 beta-lactamase in imidazole/MPD 60 minute avibactam complex | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | 著者 | Smith, C.A, Vakulenko, S.B. | | 登録日 | 2021-04-06 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | 登録日 | 2018-06-09 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | 分子名称: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-08 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6DQ6
 
 | |
7M1S
 
 | |
6DFV
 
 | | Mouse diabetogenic TCR 8F10 | | 分子名称: | 1,2-ETHANEDIOL, TCR alpha chain, TCR beta chain | | 著者 | Wang, Y, Dai, S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
7MLK
 
 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | 分子名称: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | 登録日 | 2021-04-28 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
7MHA
 
 | |
2YMO
 
 | | Crystal structure of Pf12 tandem 6-cys domains from Plasmodium falciparum | | 分子名称: | PF12 | | 著者 | Tonkin, M.L, Arredondo, S.A, Loveless, B.C, Grigg, M.E, Miller, L.H, Boulanger, M.J. | | 登録日 | 2012-10-09 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Biochemical Characterization of Plasmodium Falciparum 12 (Pf12) Reveals a Unique Inter-Domain Organization and the Potential for an Antiparallel Arrangement with Pf41
J.Biol.Chem., 288, 2013
|
|
7MRI
 
 | |
8BE4
 
 | |
8BE5
 
 | | Crystal structure of SOS1-KRasG12V-Nanobody22-Nanobody75 | | 分子名称: | Isoform 2B of GTPase KRas, Nanobody22, Nanobody75, ... | | 著者 | Fischer, B, Wohlkonig, A, Steyaert, J. | | 登録日 | 2022-10-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
8BE2
 
 | |
7MVZ
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96-Nup145N complex (Nup188 residues 1-1858; Nic96 residues 240-301; Nup145N residues 640-732) | | 分子名称: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP188 | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-05-15 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
4ZZL
 
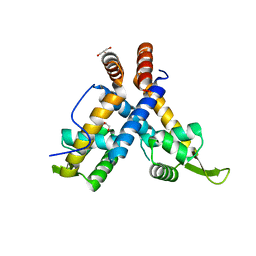 | | MexR R21W derepressor mutant causing multidrug resistance in P. aeruginosa by mexAB-oprM efflux pump expression | | 分子名称: | GLYCEROL, MULTIDRUG RESISTANCE OPERON REPRESSOR | | 著者 | Anandapadamanaban, M, Pilstal, R, Ziauddin, J.M.E, Moche, M, Wallner, B, Sunnerhagen, M. | | 登録日 | 2015-04-10 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Mutation-Induced Population Shift in the Mexr Conformational Ensemble Disengages DNA Binding: A Novel Mechanism for Marr Family Derepression.
Structure, 24, 2016
|
|
2YBO
 
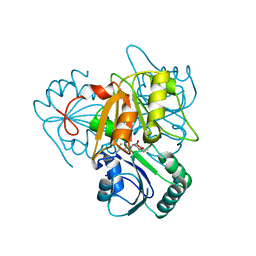 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH | | 分子名称: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | 登録日 | 2011-03-08 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
5AEX
 
 | |
2YOQ
 
 | | Structure of FAM3B PANDER E30 construct | | 分子名称: | GLYCEROL, PROTEIN FAM3B | | 著者 | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | 登録日 | 2012-10-26 | | 公開日 | 2013-01-30 | | 最終更新日 | 2013-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
4V31
 
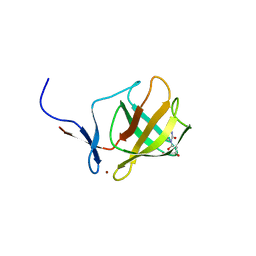 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | 分子名称: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | 著者 | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2014-10-15 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
6D9F
 
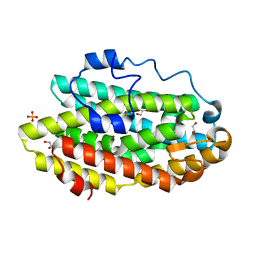 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | 著者 | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | 登録日 | 2018-04-28 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
2YXE
 
 | |
5A78
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA not cleaved | | 分子名称: | 24MER DNA, 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP *CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', DNA ENDONUCLEASE I-CVUI, ... | | 著者 | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | 登録日 | 2015-07-03 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
2YXJ
 
 | | Crystal structure of Bcl-xL in complex with ABT-737 | | 分子名称: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, Apoptosis regulator Bcl-X, CHLORIDE ION, ... | | 著者 | Czabotar, P.E, Lee, E.F, Smith, B.J, Deshayes, K, Zobel, K, Fairlie, W.D, Colman, P.M. | | 登録日 | 2007-04-26 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family
Cell Death Differ., 14, 2007
|
|
6DO7
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | 登録日 | 2018-06-09 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
