4W8C
 
 | |
2G6X
 
 | |
3UZV
 
 | | Crystal structure of the dengue virus serotype 2 envelope protein domain III in complex with the variable domains of Mab 4E11 | | 分子名称: | ETHANOL, anti-dengue Mab 4E11, envelope protein | | 著者 | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | 登録日 | 2011-12-07 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UV3
 
 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | 分子名称: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | 著者 | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | 登録日 | 2011-11-29 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | 分子名称: | Endolysin,Rhodopsin,S-arrestin | | 著者 | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | 登録日 | 2015-08-28 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (7.7 Å) | | 主引用文献 | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
4W8Y
 
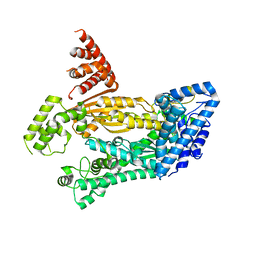 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | 分子名称: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4OZC
 
 | | Backbone Modifications in the Protein GB1 Helix and Loops: beta-ACPC21, beta-ACPC24, beta-3-Lys28, beta-3-Lys31, beta-ACPC35, beta-ACPC40 | | 分子名称: | GLYCEROL, SULFATE ION, Streptococcal Protein GB1 Backbone Modified Variant: beta-ACPC21, ... | | 著者 | Reinert, Z.E, Horne, W.S. | | 登録日 | 2014-02-14 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
6QMR
 
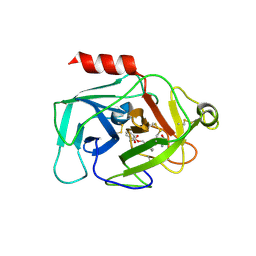 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | 分子名称: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | 著者 | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
3UFD
 
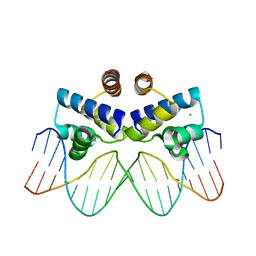 | | C.Esp1396I bound to its highest affinity operator site OM | | 分子名称: | CHLORIDE ION, DNA (5'-D(*AP*TP*GP*TP*AP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*TP*AP*CP*A)-3'), ... | | 著者 | Ball, N.J, McGeehan, J.E, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | 登録日 | 2011-11-01 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Nucleic Acids Res., 40, 2012
|
|
4WBH
 
 | |
2G6Y
 
 | |
6QOZ
 
 | | CryoEM reconstruction of Cowpea Mosaic Virus (CPMV) bound to an Affimer reagent | | 分子名称: | Affimer binding protein, Cowpea mosaic virus large subunit, RNA2 polyprotein | | 著者 | Hesketh, E.L, Tiede, C, Adamson, H, Adams, T.L, Byrne, M.J, Meshcheriakova, Y, Lomonossoff, G.P, Kruse, I, McPherson, M.J, Tomlinson, D.C, Ranson, N.A. | | 登録日 | 2019-02-13 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Affimer reagents as tools in diagnosing plant virus diseases.
Sci Rep, 9, 2019
|
|
3UFJ
 
 | | Human Thymine DNA Glycosylase Bound to Substrate Analog 2'-fluoro-2'-deoxyuridine | | 分子名称: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*GP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(UF2)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Maiti, A, Drohat, A.C. | | 登録日 | 2011-11-01 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.967 Å) | | 主引用文献 | Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DIQ
 
 | | Crystal structure of human FPPS in complex with salicylic acid derivative 3a | | 分子名称: | 2-(naphthalen-1-ylmethoxy)benzoic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | 著者 | Rondeau, J.M, Bourgier, E, Lehmann, S. | | 登録日 | 2015-09-01 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Novel Allosteric Non-Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthase by Integrated Lead Finding.
Chemmedchem, 10, 2015
|
|
4WD0
 
 | | Crystal structure of HisAp form Arthrobacter aurescens | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-05 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of HisAp form Arthrobacter aurescens
To Be Published
|
|
4V6O
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4a of the six classes) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | 登録日 | 2011-12-07 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (14.7 Å) | | 主引用文献 | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DTC
 
 | | UBL Structure | | 分子名称: | Ribosome biogenesis protein YTM1 | | 著者 | Romes, E.M, Stanley, R.E. | | 登録日 | 2015-09-17 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of the Ubiquitin-like Domain of Ribosome Assembly Factor Ytm1 and Characterization of Its Interaction with the AAA-ATPase Midasin.
J.Biol.Chem., 291, 2016
|
|
3URK
 
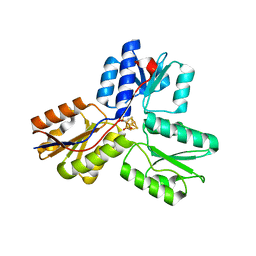 | | IspH in complex with propynyl diphosphate (1061) | | 分子名称: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, prop-2-yn-1-yl trihydrogen diphosphate | | 著者 | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | 登録日 | 2011-11-22 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UO6
 
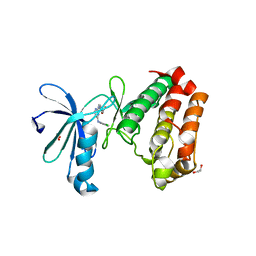 | | Aurora A in complex with YL5-083 | | 分子名称: | 1,2-ETHANEDIOL, 4-({4-[(2-chlorophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | 著者 | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2011-11-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8002 Å) | | 主引用文献 | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
4ODB
 
 | |
3UOJ
 
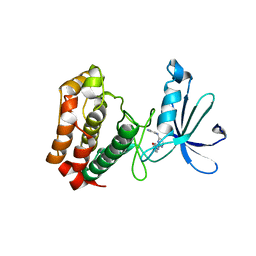 | | Aurora A in complex with RPM1715 | | 分子名称: | 4-({4-[(2-cyanophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | 著者 | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2011-11-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9003 Å) | | 主引用文献 | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
5DLV
 
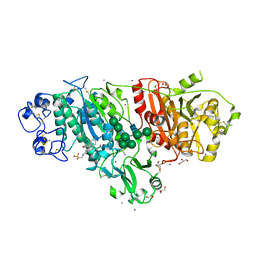 | | Crystal structure of Autotaxin (ENPP2) with tauroursodeoxycholic acid (TUDCA) | | 分子名称: | 2-{[(3alpha,5beta,7alpha,8alpha,14beta,17alpha)-3,7-dihydroxy-24-oxocholan-24-yl]amino}ethanesulfonic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | 著者 | Keune, W.J, Heidebrecht, T, von Castelmur, E, Joosten, R.P, Perrakis, A. | | 登録日 | 2015-09-07 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling.
Nat Commun, 7, 2016
|
|
5DJR
 
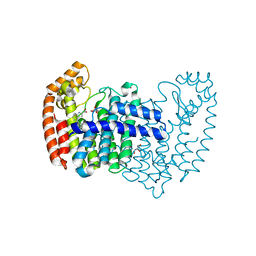 | | Crystal structure of human FPPS in complex with biaryl compound 6 | | 分子名称: | 1H,1'H-4,4'-biindole-2-carboxylic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | 著者 | Rondeau, J.M, Bourgier, E, Lehmann, S. | | 登録日 | 2015-09-02 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of Novel Allosteric Non-Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthase by Integrated Lead Finding.
Chemmedchem, 10, 2015
|
|
5E8N
 
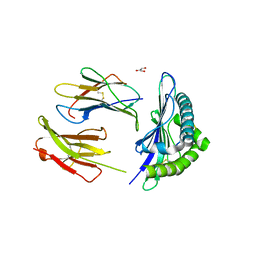 | | The structure of the TEIPP associated Trh4 peptide in complex with H-2D(b) | | 分子名称: | Beta-2-microglobulin, Ceramide synthase 5, GLYCEROL, ... | | 著者 | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | 登録日 | 2015-10-14 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
6QT9
 
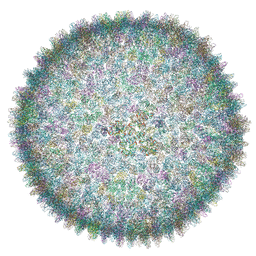 | | Cryo-EM structure of SH1 full particle. | | 分子名称: | ORF 24, ORF 25, ORF 31, ... | | 著者 | De Colibus, L, Roine, E, Walter, T.S, Ilca, S.L, Wang, X, Wang, N, Roseman, A.M, Bamford, D, Huiskonen, J.T, Stuart, D.I. | | 登録日 | 2019-02-22 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Nat Commun, 10, 2019
|
|
