6SI3
 
 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9301 | | 分子名称: | 1,2-ETHANEDIOL, 1-[7-bromanyl-9-[2,2,2-tris(fluoranyl)ethyl]carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, ... | | 著者 | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-08 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
6CBN
 
 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 7.5 | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Neamine transaminase NeoN | | 著者 | Thoden, J.B, Dow, G.T, Holden, H.M. | | 登録日 | 2018-02-03 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
7CYS
 
 | | Crystal structure of barley agmatine coumaroyltransferase (HvACT), an N-acyltransferase in BAHD superfamily | | 分子名称: | Agmatine coumaroyltransferase-1 | | 著者 | Yamane, M, Takenoya, M, Sue, M, Yajima, S. | | 登録日 | 2020-09-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure of barley agmatine coumaroyltransferase, an N-acyltransferase from the BAHD superfamily.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7GJ4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-5d232de5-1 (Mpro-P0160) | | 分子名称: | (4R)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.128 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHS
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ERI-UCB-d6de1f3c-2 (Mpro-P0018) | | 分子名称: | 1-(3-chlorophenyl)-4-(isoquinoline-4-carbonyl)piperazin-2-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7DA5
 
 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | 分子名称: | Basigin, Monocarboxylate transporter 1 | | 著者 | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | 登録日 | 2020-10-14 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
1BIR
 
 | | RIBONUCLEASE T1, PHE 100 TO ALA MUTANT COMPLEXED WITH 2' GMP | | 分子名称: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | 著者 | Doumen, J, Gonciarz, M, Zegers, I, Loris, R, Wyns, L, Steyaert, J. | | 登録日 | 1996-01-04 | | 公開日 | 1996-08-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A catalytic function for the structurally conserved residue Phe 100 of ribonuclease T1.
Protein Sci., 5, 1996
|
|
5BOW
 
 | |
3FRS
 
 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | 分子名称: | GLYCEROL, Uncharacterized protein KIAA0174 | | 著者 | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | 登録日 | 2009-01-08 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6MME
 
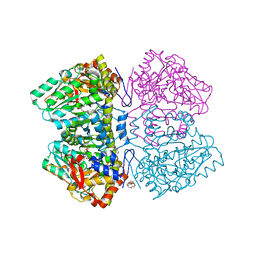 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-09-30 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
5EIS
 
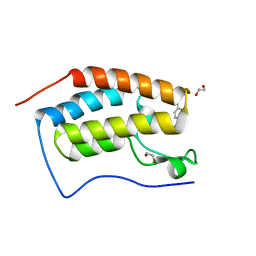 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR 3-(4-Chlorobenzyl)-7-ethyl-3,7-dihydropurine-2,6-dione | | 分子名称: | 1,2-ETHANEDIOL, 3-[(4-chlorophenyl)methyl]-7-ethyl-purine-2,6-dione, Bromodomain-containing protein 4 | | 著者 | Raux, B, Rebuffet, E, Betzi, S, Morelli, X. | | 登録日 | 2015-10-30 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Exploring Selective Inhibition of the First Bromodomain of the Human Bromodomain and Extra-terminal Domain (BET) Proteins.
J.Med.Chem., 59, 2016
|
|
5Z9X
 
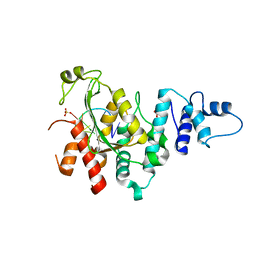 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | 分子名称: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | 著者 | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | 登録日 | 2018-02-05 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
3FWQ
 
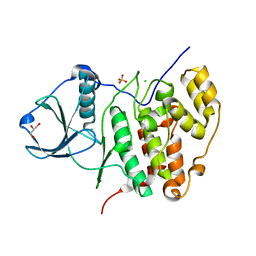 | | Inactive conformation of human protein kinase CK2 catalytic subunit | | 分子名称: | CHLORIDE ION, Casein kinase II subunit alpha, GLYCEROL, ... | | 著者 | Niefind, K, Raaf, J, Issinger, O.G. | | 登録日 | 2009-01-19 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | First inactive conformation of CK2 alpha, the catalytic subunit of protein kinase CK2
J.Mol.Biol., 386, 2009
|
|
6CMN
 
 | |
6CMR
 
 | | Closed structure of active SHP2 mutant E76D bound to SHP099 inhibitor | | 分子名称: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | 登録日 | 2018-03-06 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
5EIW
 
 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYL METHIONINE AND FRAGMENT NB3C2 | | 分子名称: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, NS5 methyltransferase, S-ADENOSYLMETHIONINE | | 著者 | Barral, K, Bricogne, G, Sharff, A. | | 登録日 | 2015-10-30 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.611 Å) | | 主引用文献 | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | 分子名称: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | 著者 | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | 登録日 | 2019-11-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
4M7B
 
 | | Human tankyrase 2 - catalytic Parp domain in complex with an inhibitor UPF1854 | | 分子名称: | 4-{2-[(6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-yl)amino]ethyl}phenol, SULFATE ION, Tankyrase-2, ... | | 著者 | Karlberg, T, Camaioni, E, Schuler, H. | | 登録日 | 2013-08-12 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Design, Synthesis, Crystallographic Studies, and Preliminary Biological Appraisal of New Substituted Triazolo[4,3-b]pyridazin-8-amine Derivatives as Tankyrase Inhibitors.
J.Med.Chem., 57, 2014
|
|
5BT4
 
 | | Crystal structure of BRD4 first bromodomain in complex with SGC-CBP30 chemical probe | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | 著者 | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2015-06-02 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of BRD4 first bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5I94
 
 | | Crystal structure of human glutaminase C in complex with the inhibitor UPGL-00019 | | 分子名称: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | 著者 | Huang, Q, Cerione, R. | | 登録日 | 2016-02-19 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.983 Å) | | 主引用文献 | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5VNS
 
 | | M.tb Antigen 85C Acyl-Enzyme Intermediate with Tetrahydrolipstatin | | 分子名称: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ... | | 著者 | Goins, C.M, Ronning, D.R. | | 登録日 | 2017-05-01 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Mycolyltransferase fromMycobacterium tuberculosisin covalent complex with tetrahydrolipstatin provides insights into antigen 85 catalysis.
J. Biol. Chem., 293, 2018
|
|
5IQ6
 
 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | 分子名称: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | 著者 | Tarantino, D, Mastrangelo, E, Milani, M. | | 登録日 | 2016-03-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
1DK9
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | 分子名称: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | 著者 | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | 登録日 | 1999-12-06 | | 公開日 | 1999-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
4O8Y
 
 | | Zinc-free Rpn11 in complex with Rpn8 | | 分子名称: | 1,2-ETHANEDIOL, 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8 | | 著者 | Worden, E.J, Padovani, C, Martin, A. | | 登録日 | 2013-12-30 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | 分子名称: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-11 | | 公開日 | 2015-12-23 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
