2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | 分子名称: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | 著者 | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | 登録日 | 2010-11-01 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2GPV
 
 | | Estrogen Related Receptor-gamma ligand binding domain complexed with 4-hydroxy-tamoxifen and a SMRT peptide | | 分子名称: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma, Nuclear receptor corepressor 2 | | 著者 | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | 登録日 | 2006-04-18 | | 公開日 | 2006-09-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
2RT5
 
 | |
5X8X
 
 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Compound A. | | 分子名称: | (3R,4R)-4-[4-cyclopropyl-5-[3-(2-methylpropyl)cyclobutyl]-1,2,4-triazol-3-yl]-N-(2,4-dimethylphenyl)-1-ethanoyl-pyrrolidine-3-carboxamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | 著者 | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | 登録日 | 2017-03-03 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
8G8F
 
 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
5OYB
 
 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | 分子名称: | Anoctamin-1, CALCIUM ION | | 著者 | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | 登録日 | 2017-09-08 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYG
 
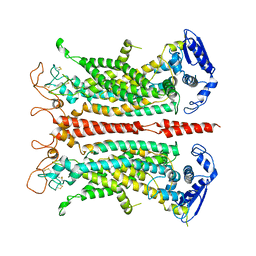 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | 分子名称: | Anoctamin-1 | | 著者 | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | 登録日 | 2017-09-08 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
7A4A
 
 | |
5N8O
 
 | |
6MZX
 
 | |
6MZY
 
 | |
6YGI
 
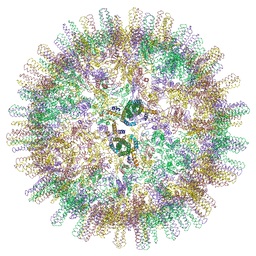 | |
6YGH
 
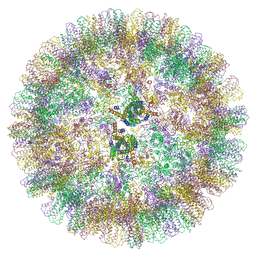 | | Duck hepatitis B virus capsid | | 分子名称: | Capsid protein | | 著者 | Makbul, C, Bottcher, B. | | 登録日 | 2020-03-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Slowly folding surface extension in the prototypic avian hepatitis B virus capsid governs stability.
Elife, 9, 2020
|
|
6GCT
 
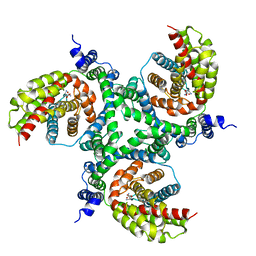 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | 分子名称: | GLUTAMINE, Neutral amino acid transporter B(0) | | 著者 | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | 登録日 | 2018-04-19 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | 分子名称: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | 著者 | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | 登録日 | 2018-02-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
6H3C
 
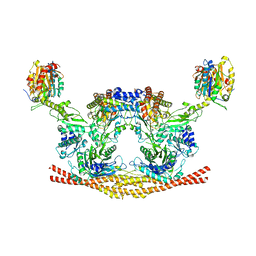 | | Cryo-EM structure of the BRISC complex bound to SHMT2 | | 分子名称: | BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, BRISC complex subunit Abraxas 2, ... | | 著者 | Bunker, R.D, Rabl, J, Thoma, N.H. | | 登録日 | 2018-07-18 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
6K3I
 
 | |
6KMF
 
 | | FimA type V pilus from P.gingivalis | | 分子名称: | Major fimbrium subunit FimA type-1 | | 著者 | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | 登録日 | 2021-05-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.35534 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
