4O76
 
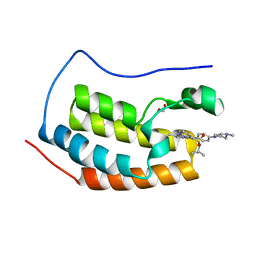 | | Crystal structure of the first bromodomain of human BRD4 in complex with TG101209 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide | | 著者 | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | 登録日 | 2013-12-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4OBD
 
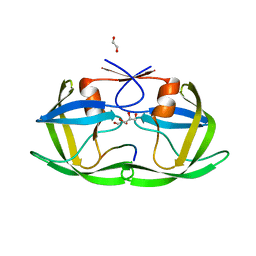 | |
3GI6
 
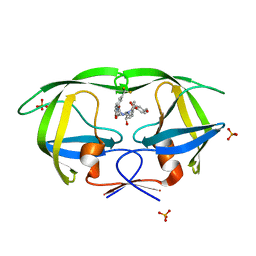 | |
5YWC
 
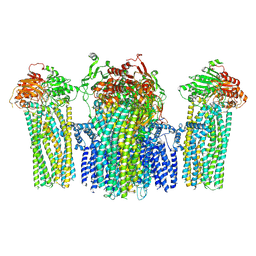 | |
7FI4
 
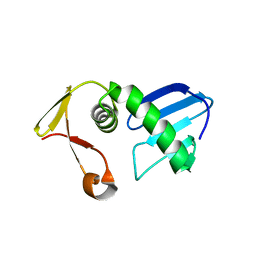 | | Structure of AcrIF13 | | 分子名称: | AcrIF13 | | 著者 | Feng, Y, Gao, T. | | 登録日 | 2021-07-30 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
3FUR
 
 | | Crystal Structure of PPARg in complex with INT131 | | 分子名称: | 2,4-dichloro-N-[3,5-dichloro-4-(quinolin-3-yloxy)phenyl]benzenesulfonamide, CHLORIDE ION, Nuclear receptor coactivator 1, ... | | 著者 | Wang, Z, Liu, J, Walker, N. | | 登録日 | 2009-01-14 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | INT131: a selective modulator of PPAR gamma.
J.Mol.Biol., 386, 2009
|
|
4OBK
 
 | |
6DGT
 
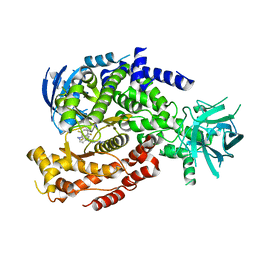 | | Selective PI3K beta inhibitor bound to PI3K delta | | 分子名称: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Somoza, J, Villasenor, A, McGrath, M. | | 登録日 | 2018-05-18 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
3FVL
 
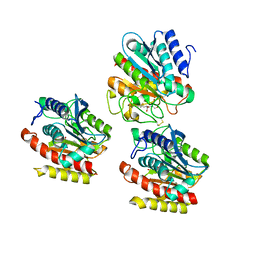 | | Crystallogic studies on the Complex of Carboxypeptidase A with inhibitors using alpha-hydroxy ketone as zinc-binding group | | 分子名称: | (2R)-2-benzyl-5-hydroxy-4-oxopentanoic acid, Carboxypeptidase A1, ZINC ION | | 著者 | Wang, S.F, Jin, J.-Y, Tian, G.R. | | 登録日 | 2009-01-16 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Optical 2-benzyl-5-hydroxy4oxopentanoic acids against carboxypeptidase A: Synthesis, kinetic evaluation and X-ray crystallographic study
CHIN.CHEM.LETT., 21, 2010
|
|
5YW9
 
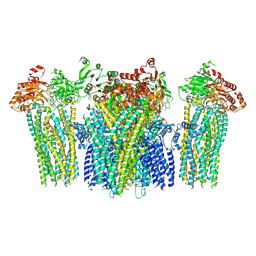 | |
5YWB
 
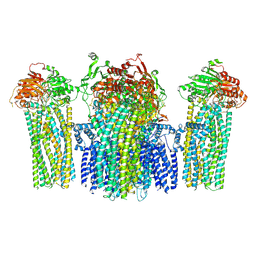 | |
6DNE
 
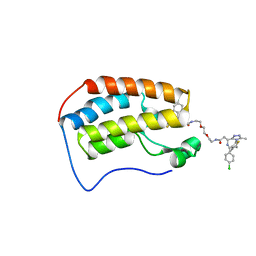 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | 分子名称: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | 著者 | Ren, C, Zhou, M.M. | | 登録日 | 2018-06-06 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.958 Å) | | 主引用文献 | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZFM
 
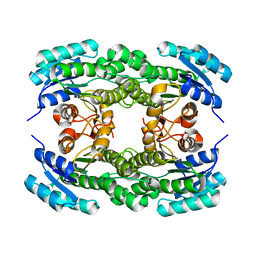 | | Ketoreductase LbCR mutant - M6 | | 分子名称: | 3-oxoacyl-acyl carrier protein reductase | | 著者 | Gong, X.M, Zheng, G.W, Xu, J.H. | | 登録日 | 2018-03-06 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
8FPE
 
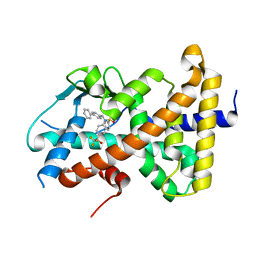 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | 分子名称: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | 登録日 | 2023-01-04 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5ZGU
 
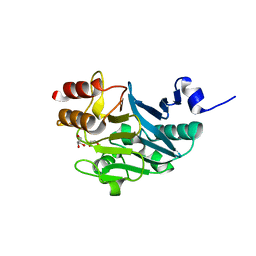 | |
5CW6
 
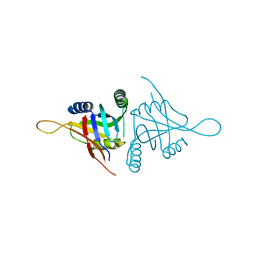 | |
5ZI0
 
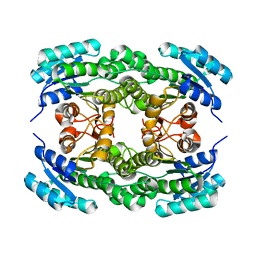 | | Ketoreductase LbCR mutant - M8 | | 分子名称: | 3-oxoacyl-acyl carrier protein reductase | | 著者 | Gong, X.M, Zheng, G.W, Xu, J.H. | | 登録日 | 2018-03-14 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
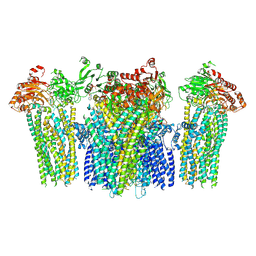
5YW8
 
 | |
7FH0
 
 | |
7EZT
 
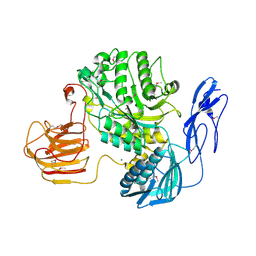 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | 分子名称: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | 著者 | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | 登録日 | 2021-06-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | 著者 | Cao, Y.L, Gu, D.D, Gao, S. | | 登録日 | 2021-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55090284 Å) | | 主引用文献 | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | 分子名称: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | 著者 | Shi, Y, Huang, G, Zhan, X. | | 登録日 | 2021-07-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
5Z96
 
 | | Structure of the mouse TRPC4 ion channel | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Duan, J, Li, Z, Li, J, Zhang, J. | | 登録日 | 2018-02-02 | | 公開日 | 2018-04-18 | | 最終更新日 | 2018-08-29 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structure of the mouse TRPC4 ion channel.
Nat Commun, 9, 2018
|
|
6QEI
 
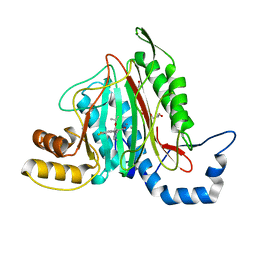 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 5,6-Difluoro-3-(2-isopropoxy-4-piperazin-1-yl-phenyl)-1H-indole-2-carboxylic acid amide | | 分子名称: | 1,2-ETHANEDIOL, 5,6-bis(fluoranyl)-3-(4-piperazin-1-yl-2-propan-2-yloxy-phenyl)-1~{H}-indole-2-carboxamide, DIMETHYL SULFOXIDE, ... | | 著者 | Musil, D, Heinrich, T, Lehmann, M. | | 登録日 | 2019-01-07 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
8FO5
 
 | |
