9AT6
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
6FGT
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 3 | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | 著者 | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | 登録日 | 2018-01-11 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
1MNG
 
 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | 分子名称: | AZIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | 著者 | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | 登録日 | 1994-07-13 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
8S69
 
 | | Low pH (5.5) as-isolated MSOX movie series dataset 40 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [22.8 MGy] | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | 著者 | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
4ZYA
 
 | | The N-terminal extension domain of human asparaginyl-tRNA synthetase | | 分子名称: | Asparagine--tRNA ligase, cytoplasmic, CHLORIDE ION, ... | | 著者 | Park, J.S, Park, M.C, Goughnour, P, Kim, H.S, Kim, S.J, Kim, H.J, Kim, S.H, Han, B.W. | | 登録日 | 2015-05-21 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Unique N-terminal extension domain of human asparaginyl-tRNA synthetase elicits CCR3-mediated chemokine activity.
Int. J. Biol. Macromol., 120, 2018
|
|
8S64
 
 | | Low pH (5.5) as-isolated MSOX movie series dataset 10 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [5.7 MGy] | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | 著者 | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RWL
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | 著者 | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | 登録日 | 2024-02-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Allostery and Evolution: A Molecular Journey Through the Structural and Dynamical Landscape of an Enzyme Super Family.
Mol.Biol.Evol., 42, 2025
|
|
7UMY
 
 | | Crystal structure of Acinetobacter baumannii FabI in complex with NAD and Fabimycin ((S,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide) | | 分子名称: | (2E)-3-[(7S)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hajian, B. | | 登録日 | 2022-04-08 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
7UQO
 
 | | Pathogenesis related 10-10 (S)-tetrahydropapaverine complex | | 分子名称: | (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pathogenesis Related 10-10 protein | | 著者 | Carr, S.C, Ng, K.K.S. | | 登録日 | 2022-04-19 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
5SVJ
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the closed, apo state | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.984 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
6U3A
 
 | |
6FAE
 
 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | 分子名称: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | 著者 | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | 登録日 | 2017-12-15 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
9H8P
 
 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, mutant DTT | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Rozeboom, H.J, Fraaije, M.W. | | 登録日 | 2024-10-29 | | 公開日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Biocatalytic Cleavage of para-Acetoxy Benzyl Ethers: Application to Protecting Group Chemistry
Acs Catalysis, 15, 2025
|
|
8IJM
 
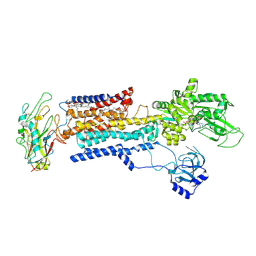 | | Cyo-EM structure of K794A non-gastric proton pump in Na+ bound E1AMPPCP state | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Abe, K. | | 登録日 | 2023-02-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | An unusual conformation from Na + -sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na + -binding.
Biochim Biophys Acta Mol Cell Res, 1870, 2023
|
|
4RH1
 
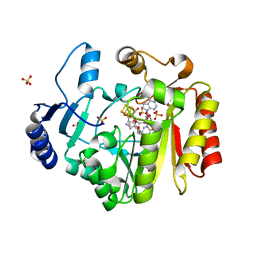 | | Spore photoproduct lyase C140A/S76C mutant with bound AdoMet and dinucleoside spore photoproduct | | 分子名称: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, SULFATE ION, ... | | 著者 | Benjdia, A, Heil, K, Winkler, A, Carell, T, Schlichting, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase.
Chem.Commun.(Camb.), 50, 2014
|
|
9EIA
 
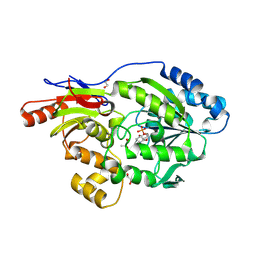 | | A broad-substrate spectrum lactate racemase A from Isosphaera pallida in complex with copurified D-lactate | | 分子名称: | 1,2-ETHANEDIOL, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, A broad-substrate spectrum lactate racemase A, ... | | 著者 | Gatreddi, S, Hausinger, R.P, Hu, J. | | 登録日 | 2024-11-25 | | 公開日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural Basis for Catalysis and Substrate Specificity of a LarA Racemase with a Broad Substrate Spectrum.
Biorxiv, 2024
|
|
6FGV
 
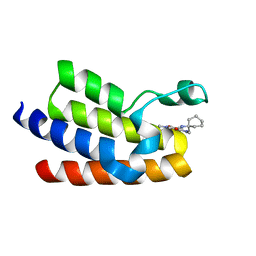 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 3 | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | 著者 | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | 登録日 | 2018-01-11 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
8RW8
 
 | |
6FIW
 
 | |
5TB5
 
 | | Crystal structure of full-length farnesylated and methylated KRAS4b in complex with PDE-delta (crystal form I - with partially disordered hypervariable region) | | 分子名称: | 1,2-ETHANEDIOL, FARNESYL, GTPase KRas, ... | | 著者 | Dharmaiah, S, Tran, T.H, Simanshu, D.K. | | 登録日 | 2016-09-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of recognition of farnesylated and methylated KRAS4b by PDE delta.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9B4T
 
 | | Crystal structure of the MRAS-p110alpha complex | | 分子名称: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Czyzyk, D.J, Simanshu, D.K. | | 登録日 | 2024-03-21 | | 公開日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural insights into isoform-specific RAS-PI3K alpha interactions and the role of RAS in PI3K alpha activation.
Nat Commun, 16, 2025
|
|
6FBC
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification at the 3'-terminus of the primer. | | 分子名称: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | 著者 | Kropp, H.M, Diederichs, K, Marx, A. | | 登録日 | 2017-12-19 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6R3V
 
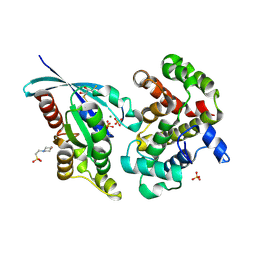 | | Crystal Structure of RhoA-GDP-Pi in Complex with RhoGAP | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Jin, Y. | | 登録日 | 2019-03-21 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A GAP-GTPase-GDP-PiIntermediate Crystal Structure Analyzed by DFT Shows GTP Hydrolysis Involves Serial Proton Transfers.
Chemistry, 25, 2019
|
|
6HWH
 
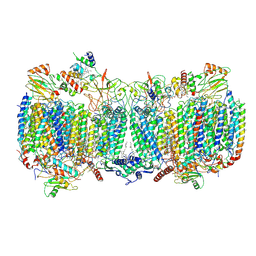 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | 分子名称: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | 著者 | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | 登録日 | 2018-10-12 | | 公開日 | 2018-11-07 | | 最終更新日 | 2025-10-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8Q1X
 
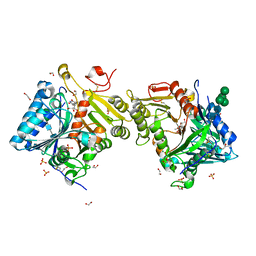 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | 著者 | Roske, Y, Daumke, O, Damme, M. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
