7UQ6
 
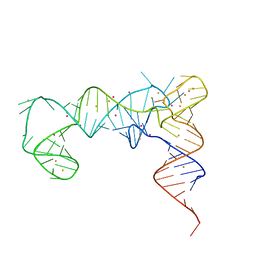 | | tRNA T-box antiterminator fusion, construct #4 | | 分子名称: | BARIUM ION, POTASSIUM ION, RNA (86-MER) | | 著者 | Grigg, J.C, Price, I.R, Ke, A. | | 登録日 | 2022-04-19 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch
Crystals, 12, 2022
|
|
2O20
 
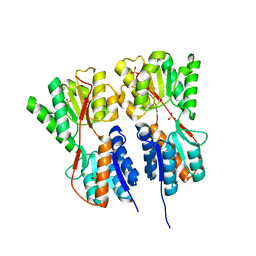 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | 分子名称: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | 著者 | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | 登録日 | 2006-11-29 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
2JZZ
 
 | |
7BLV
 
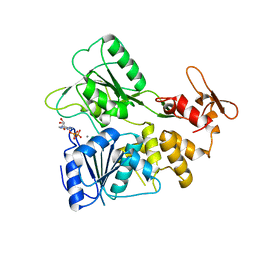 | |
5LW4
 
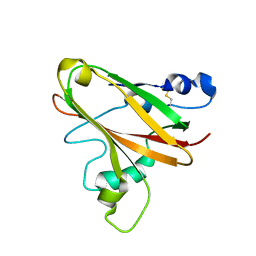 | |
5LCB
 
 | | In situ atomic-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum obtained combining solid-state NMR spectroscopy, cryo electron microscopy and polarization spectroscopy | | 分子名称: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll c-binding protein | | 著者 | Nielsen, J.T, Kulminskaya, N.V, Bjerring, M, Linnanto, J.M, Ratsep, M, Pedersen, M, Lambrev, P.H, Dorogi, M, Garab, G, Thomsen, K, Jegerschold, C, Frigaard, N.U, Lindahl, M, Nielsen, N.C. | | 登録日 | 2016-06-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (26.5 Å), SOLID-STATE NMR | | 主引用文献 | In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum.
Nat Commun, 7, 2016
|
|
7R25
 
 | | Bacillus pumilus Lipase A | | 分子名称: | CITRIC ACID, Lipase, PHOSPHATE ION, ... | | 著者 | Lund, B.A. | | 登録日 | 2022-02-04 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.87 Å) | | 主引用文献 | Structure and Mechanism of a Cold-Adapted Bacterial Lipase
Biochemistry, 61, 2022
|
|
7D74
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | 著者 | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | 登録日 | 2020-10-02 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-12-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | 分子名称: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | 著者 | Zhao, H, Dai, Y, Lee, C.H. | | 登録日 | 2022-04-01 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7BO3
 
 | | Human Butyrylcholinesterase in complex with N-(2-(1H-Indol-3-yl)ethyl)-2-cycloheptyl-N-methylethan-1-amine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Meden, A, Knez, D, Nachon, F, Gobec, S. | | 登録日 | 2021-01-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | From tryptophan-based amides to tertiary amines: Optimization of a butyrylcholinesterase inhibitor series.
Eur.J.Med.Chem., 234, 2022
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | 分子名称: | Flagellar M-ring protein | | 著者 | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | 登録日 | 2020-10-07 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | 分子名称: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | 著者 | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | 登録日 | 2020-11-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
3AQU
 
 | | Crystal structure of a class V chitinase from Arabidopsis thaliana | | 分子名称: | At4g19810, CITRATE ANION | | 著者 | Numata, T, Ohnuma, T, Osawa, T, Fukamizo, T. | | 登録日 | 2010-11-19 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis
Planta, 234, 2011
|
|
7BHJ
 
 | |
7MSF
 
 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | 分子名称: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | 著者 | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | 登録日 | 1998-05-20 | | 公開日 | 1998-11-11 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
3AVR
 
 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | 著者 | Sengoku, T, Yokoyama, S. | | 登録日 | 2011-03-07 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
7RSY
 
 | | HIV-1 gp120 complex with CJF-III-049-R | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | 著者 | Liang, S, Hendrickson, W.A. | | 登録日 | 2021-08-12 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
7DJT
 
 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | 分子名称: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | 著者 | Cai, Y, Zhang, H. | | 登録日 | 2020-11-21 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
5M5J
 
 | | Thioredoxin reductase from Giardia duodenalis | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, Thioredoxin reductase | | 著者 | Fiorillo, A, Ilari, A, Lalle, M. | | 登録日 | 2016-10-21 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural characterization of Giardia duodenalis thioredoxin reductase (gTrxR) and computational analysis of its interaction with NBDHEX.
Eur J Med Chem, 135, 2017
|
|
5M5T
 
 | |
2JN0
 
 | | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A. | | 分子名称: | Hypothetical lipoprotein ygdR | | 著者 | Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-12-15 | | 公開日 | 2007-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
7DHW
 
 | | Crystal structure of myosin-XI motor domain in complex with ADP-ALF4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Haraguchi, T, Tamanaha, M, Yoshimura, K, Imi, T, Tominaga, M, Sakayama, H, Nishiyama, T, Ito, K, Murata, T. | | 登録日 | 2020-11-17 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of ultrafast myosin, its amino acid sequence, and structural features.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5M65
 
 | |
5M6J
 
 | |
