3C52
 
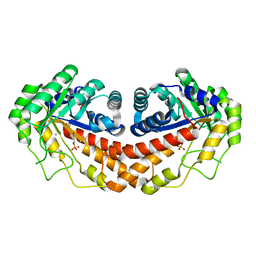 | | Class II fructose-1,6-bisphosphate aldolase from helicobacter pylori in complex with phosphoglycolohydroxamic acid, a competitive inhibitor | | 分子名称: | CALCIUM ION, Fructose-bisphosphate aldolase, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | 著者 | Coincon, M, Sygusch, J. | | 登録日 | 2008-01-30 | | 公開日 | 2008-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis and Biochemical Evaluation of Selective Inhibitors of Class II Fructose Bisphosphate Aldolases: Towards New Synthetic Antibiotics.
Chemistry, 14, 2008
|
|
7BOD
 
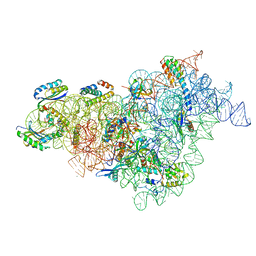 | | Bacterial 30S ribosomal subunit assembly complex state M (body domain) | | 分子名称: | 16S rRNA (body domain of 30S subunit), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | 登録日 | 2021-01-25 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BOF
 
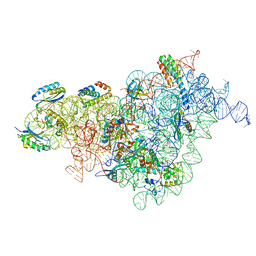 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | 分子名称: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2021-01-25 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
8DKN
 
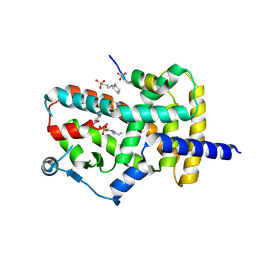 | | PPARg bound to T0070907 and Co-R peptide | | 分子名称: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | 著者 | Larsen, N.A, Tsai, J. | | 登録日 | 2022-07-05 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | 分子名称: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Larsen, N.A. | | 登録日 | 2022-07-24 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DKV
 
 | | PPARg bound to JTP-426467 and Co-R peptide | | 分子名称: | 2-chloro-N-[4-(5-methyl-1,3-benzoxazol-2-yl)phenyl]-5-nitrobenzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1, ... | | 著者 | Larsen, N.A. | | 登録日 | 2022-07-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
5E7V
 
 | | Potent Vitamin D Receptor Agonist | | 分子名称: | 1-ALPHA-HYDROXY-27-NOR-25-O-CARBONYL-VITAMIN D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Otero, R, Seoane, S, Sigueiro, R, Belorusova, A.Y, Maestro, M.A, Perez-Fernandez, R, Rochel, N, Mourino, A. | | 登録日 | 2015-10-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Carborane-based design of a potent vitamin D receptor agonist.
Chem Sci, 7, 2016
|
|
3B91
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates Ade38(2AP) and 2',5'-Phosphodiester Linkage Mutations at the Active Site | | 分子名称: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2007-11-02 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
5GT4
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol | | 分子名称: | 4-{[(1R,2S,3R,5Z,7E,14beta,17alpha)-1,3,25-trihydroxy-9,10-secocholesta-5,7,10-trien-2-yl]oxy}butanenitrile, Vitamin D3 receptor | | 著者 | Takimoto-Kamimura, M. | | 登録日 | 2016-08-18 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol
To Be Published
|
|
3BAL
 
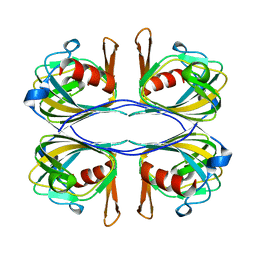 | | Crystal Structure of an Acetylacetone Dioxygenase from Acinetobacter johnsonii | | 分子名称: | Acetylacetone-cleaving enzyme, ZINC ION | | 著者 | Stranzl, G.R, Wagner, U.G, Straganz, G, Steiner, W, Kratky, C. | | 登録日 | 2007-11-08 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of an Acetylacetone Dioxygenase from Acinetobacter johnsonii
To be Published
|
|
8PJJ
 
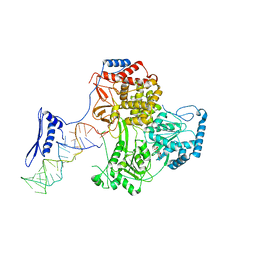 | |
5GTN
 
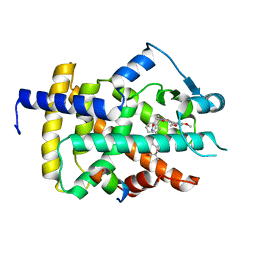 | | Human PPARgamma ligand binding dmain complexed with R35 | | 分子名称: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Suh, S.W. | | 登録日 | 2016-08-22 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
3B58
 
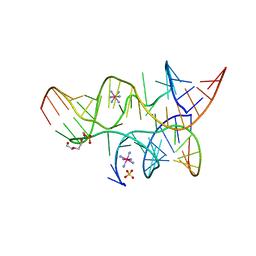 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | 分子名称: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2007-10-25 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | 分子名称: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | 著者 | Li, H, Wu, Y, Liang, H, Liu, Y. | | 登録日 | 2022-10-16 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3BA7
 
 | |
8GWO
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
3B5S
 
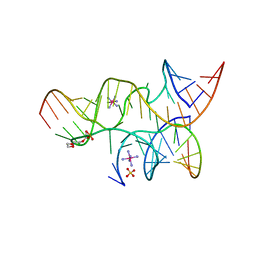 | | Minimally Hinged Hairpin Ribozyme Incorporates A38DAP Mutation and 2'-O-methyl Modification at the Active Site | | 分子名称: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2007-10-26 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
8GWB
 
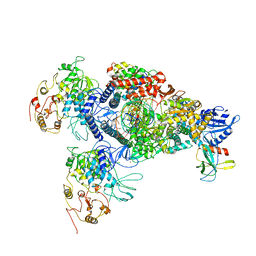 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | 分子名称: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-16 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
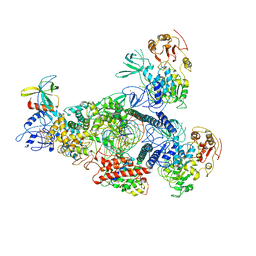 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
6FT6
 
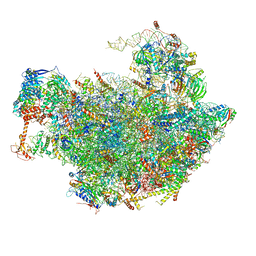 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | 分子名称: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | 著者 | Schuller, J.M, Falk, S, Conti, E. | | 登録日 | 2018-02-20 | | 公開日 | 2018-03-28 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
4O2H
 
 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | 分子名称: | protein BCAM1869 | | 著者 | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-12-17 | | 公開日 | 2014-01-22 | | 最終更新日 | 2014-10-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
4NH1
 
 | | Crystal structure of a heterotetrameric CK2 holoenzyme complex carrying the Andante-mutation in CK2beta and consistent with proposed models of autoinhibition and trans-autophosphorylation | | 分子名称: | Casein kinase II subunit alpha, Casein kinase II subunit beta, GLYCEROL, ... | | 著者 | Schnitzler, A, Issinger, O.-G, Niefind, K. | | 登録日 | 2013-11-04 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The Protein Kinase CK2(Andante) Holoenzyme Structure Supports Proposed Models of Autoregulation and Trans-Autophosphorylation
J.Mol.Biol., 426, 2014
|
|
