5W0R
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | 分子名称: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6YBJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 3e | | 分子名称: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[(2-methylpyrazol-3-yl)methoxy]phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | 著者 | Dokurno, P, Surgenor, A.E, Murray, J.B. | | 登録日 | 2020-03-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
4RAV
 
 | | Crystal structure of scFvC4 in complex with the first 17 AA of huntingtin | | 分子名称: | Huntingtin, SULFATE ION, Single-chain Fv, ... | | 著者 | De Genst, E, Chirgadze, D.Y, Dobson, C.M. | | 登録日 | 2014-09-11 | | 公開日 | 2015-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of a single-chain fv bound to the 17 N-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression.
J.Mol.Biol., 427, 2015
|
|
6N85
 
 | | Resistance to inhibitors of cholinesterase 8A (Ric8A) protein in complex with MBP-tagged transducin-alpha residues 327-350 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, Synembryn-A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Srivastava, D, Gakhar, L, Artemyev, N.O. | | 登録日 | 2018-11-28 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
6QYN
 
 | | Structure of MBP-Mcl-1 in complex with compound 10d | | 分子名称: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-09 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
3LRH
 
 | |
8VLX
 
 | |
4XAI
 
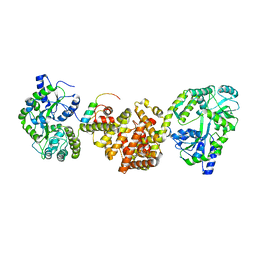 | | Crystal Structure of red flour beetle NR2E1/TLX | | 分子名称: | Grunge, isoform J, Maltose-binding periplasmic protein,Tailless ortholog, ... | | 著者 | Zhi, X, Zhou, E, Xu, E. | | 登録日 | 2014-12-14 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
6X9O
 
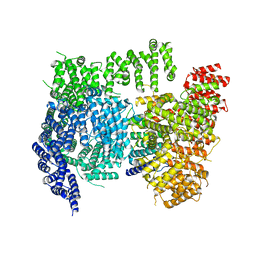 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | 分子名称: | 40-kDa huntingtin-associated protein, Huntingtin | | 著者 | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-17 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
8W15
 
 | |
6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | 分子名称: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | 著者 | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | 登録日 | 2018-05-30 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
5EDU
 
 | |
5V6Y
 
 | |
5E7U
 
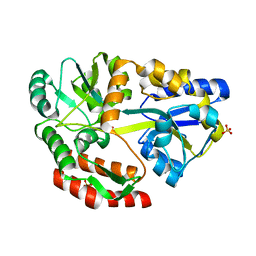 | |
4FEB
 
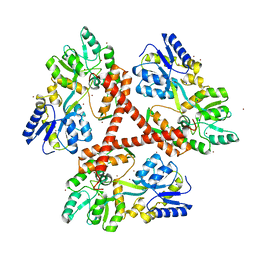 | | Crystal Structure of Htt36Q3H-EX1-X1-C2(Beta) | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, SODIUM ION, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-29 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
5JJ4
 
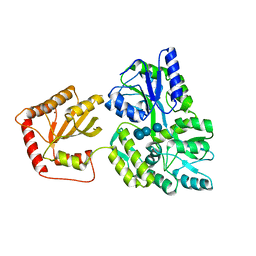 | | Crystal Structure of a Variant Human Activation-induced Deoxycytidine Deaminase as an MBP fusion protein | | 分子名称: | CALCIUM ION, Maltose-binding periplasmic protein,Single-stranded DNA cytosine deaminase, ZINC ION, ... | | 著者 | Pedersen, L.C, Goodman, M.F, Pham, P, Afif, S.A. | | 登録日 | 2016-04-22 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
DNA Repair (Amst.), 43, 2016
|
|
4FED
 
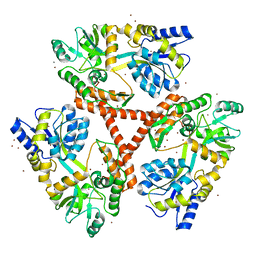 | | Crystal Structure of Htt36Q3H | | 分子名称: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
6EG3
 
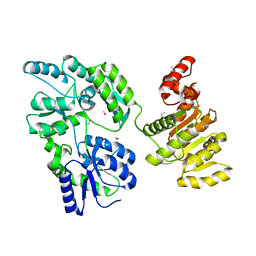 | | Crystal structure of human BRM in complex with compound 15 | | 分子名称: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | 著者 | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | 登録日 | 2018-08-17 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6DD5
 
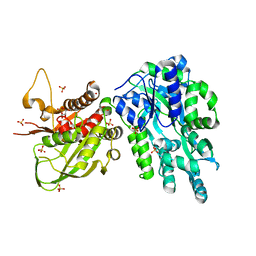 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | 分子名称: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | 著者 | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | 登録日 | 2018-05-09 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
5W0U
 
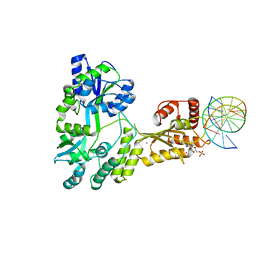 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6EG2
 
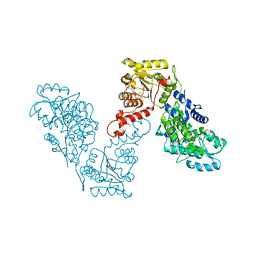 | | Crystal structure of human BRM in complex with compound 16 | | 分子名称: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | 著者 | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | 登録日 | 2018-08-17 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
5LEM
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-08-02 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
4FE8
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-29 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FEC
 
 | | Crystal Structure of Htt36Q3H | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
5LEL
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-11-15 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
