9BP9
 
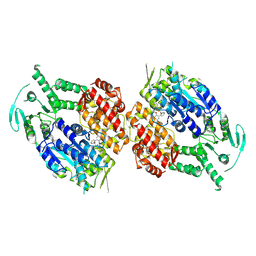 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, dimer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
9BPA
 
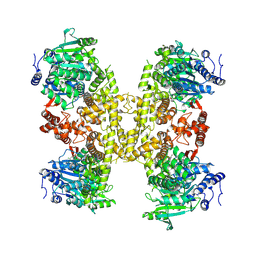 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, tetramer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
8RO1
 
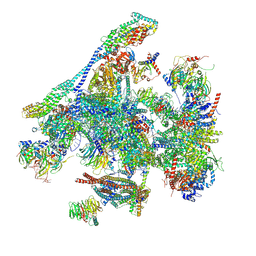 | | Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'') | | Descriptor: | CWF19-like protein 1 homolog, CWF19-like protein 2 homolog, Cell division cycle 5-like protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO0
 
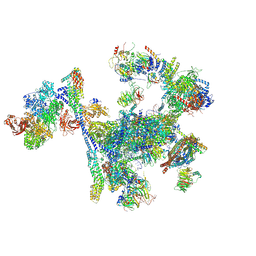 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | Descriptor: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8I0V
 
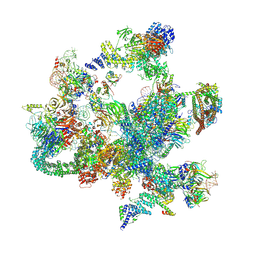 | | The cryo-EM structure of human post-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0P
 
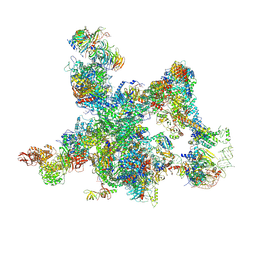 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0W
 
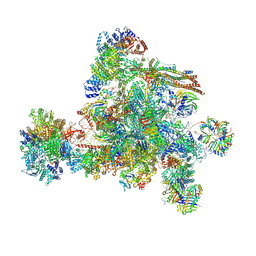 | | The cryo-EM structure of human C complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Coiled-coil domain-containing protein 94, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
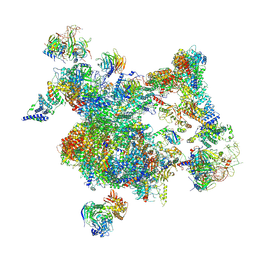 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0U
 
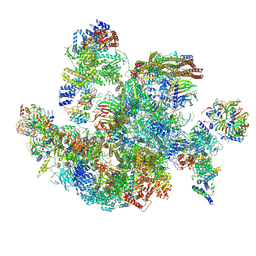 | | The cryo-EM structure of human Bact-IV complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0T
 
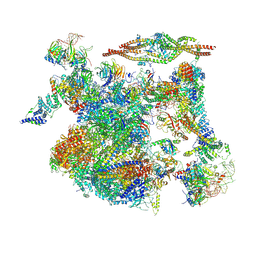 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
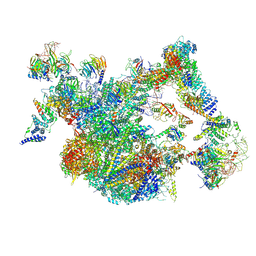 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8WTK
 
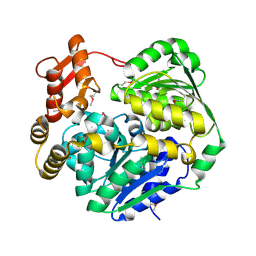 | | Crystal structure of Cas3-DExD+MG | | Descriptor: | CRISPR-associated helicase Cas3, MAGNESIUM ION | | Authors: | Mo, X. | | Deposit date: | 2023-10-18 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insight into dynamic characteristic of the polymerized forms of kinetoplastid membrane protein-11
To be published
|
|
9FMD
 
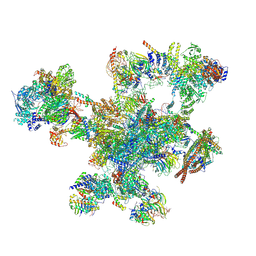 | |
9ASK
 
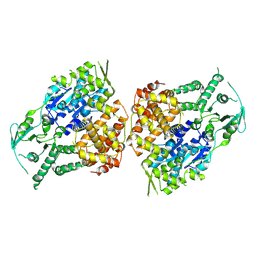 | |
9C5Q
 
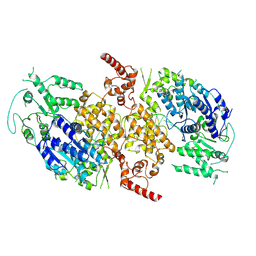 | |
9ASL
 
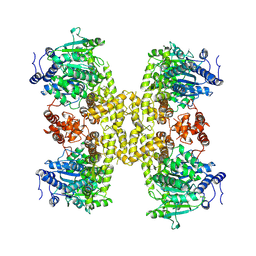 | |
9ASJ
 
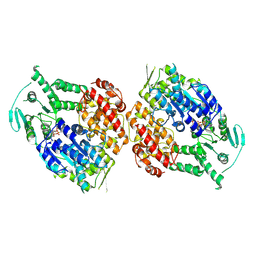 | |
8W0A
 
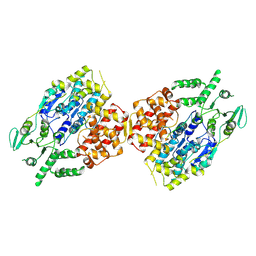 | |
8SSW
 
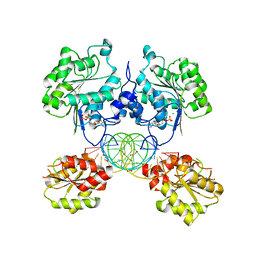 | |
8QPA
 
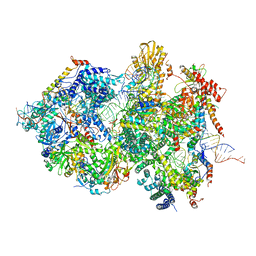 | | Cryo-EM Structure of Pre-B+5'ssLNG Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPB
 
 | | Cryo-EM Structure of Pre-B+ATP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R0B
 
 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R08
 
 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QP9
 
 | | Cryo-EM Structure of Pre-B+AMPPNP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPK
 
 | | Cryo-EM Structure of Pre-B+5'ss Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
