2NOS
 
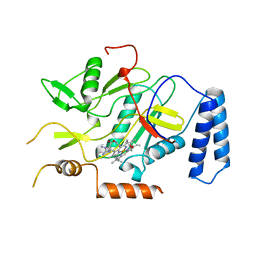 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114), AMINOGUANIDINE COMPLEX | | Descriptor: | AMINOGUANIDINE, IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
4ORB
 
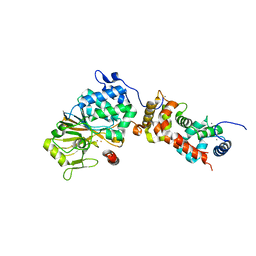 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
7NWJ
 
 | |
1PER
 
 | |
4OXY
 
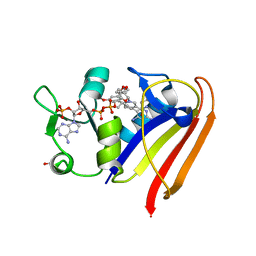
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OSG
 
 | | Klebsiella pneumoniae complexed with NADPH and 6-ethyl-5-[(3R)-3-[3-methoxyl-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Klebsiella pneumoniae Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal Features for Potency and Selectivity.
Antimicrob.Agents Chemother., 58, 2014
|
|
6W11
 
 | |
7O0Q
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 12 (ADO_AD_066) | | Descriptor: | 3-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-8-[6-(methylamino)pyrimidin-4-yl]-1,3,8-triazaspiro[4.5]decan-2-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 12 (ADO_AD_066)
To Be Published
|
|
6W12
 
 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to the Tumor-Associated Tn Antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, C-type lectin domain family 10 member A, CALCIUM ION, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
4OZI
 
 | | S2 protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HLA class II histocompatibility antigen, ... | | Authors: | Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2014-02-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6W59
 
 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC063) | | Descriptor: | 3,5-bis(fluoranyl)-~{N}-[3-[[4-(trifluoromethyloxy)phenyl]sulfamoyl]phenyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
1PG4
 
 | | Acetyl CoA Synthetase, Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, CHLORIDE ION, ... | | Authors: | Gulick, A.M, Starai, V.J, Horswill, A.R, Homick, K.M, Escalante-Semerena, J.C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.75 A Crystal Structure of Acetyl-CoA Synthetase Bound
to Adenosine-5'-propylphosphate and Coenzyme A
Biochemistry, 42, 2003
|
|
5DZG
 
 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a xyloglucan tetradecasaccharide | | Descriptor: | VvEG16, endo-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
4GDX
 
 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
3E3Q
 
 | |
2MZE
 
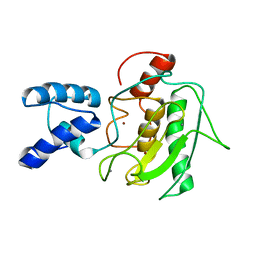 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) | | Descriptor: | CALCIUM ION, Matrilysin, ZINC ION | | Authors: | Prior, S.H, Fulcher, Y.G, Van Doren, S.R. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
4FZB
 
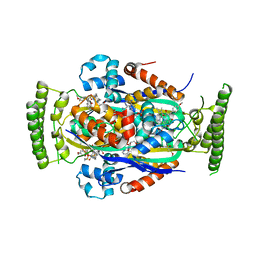 | | Structure of thymidylate synthase ThyX complexed to a new inhibitor | | Descriptor: | 2-hydroxy-3-(4-methoxybenzyl)naphthalene-1,4-dione, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Basta, T, Boum, Y, Briffotaux, J, Becker, H.F, Lamarre-Jouenne, I, Lambry, J.C, Skouloubris, S, Liebl, U, van Tilbeurgh, H, Graille, M, Myllylkallio, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic and structural basis for inhibition of thymidylate synthase ThyX.
Open Biology, 2, 2012
|
|
7NVK
 
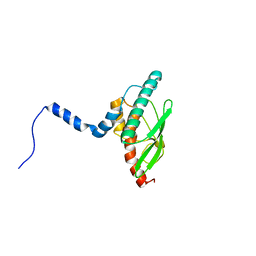 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
5AXS
 
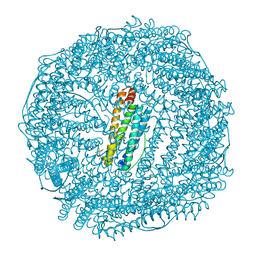 | | Crystal structure of CdCat-Fn | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Abe, S, Nakajima, H, Kondo, M, Nakane, T, Nakao, T, Ueno, T, Watanabe, Y. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Construction of an enterobactin analogue with symmetrically arranged monomer subunits of ferritin
Chem.Commun.(Camb.), 51, 2015
|
|
1PHF
 
 | |
2DAE
 
 | | Solution Structure of the N-terminal CUE Domain in the Human Mitogen-activated Protein Kinase Kinase Kinase 7 Interacting Protein 2 (MAP3K7IP2) | | Descriptor: | KIAA0733 protein | | Authors: | Zhao, C, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal CUE Domain in the Human Mitogen-activated Protein Kinase Kinase Kinase 7 Interacting Protein 2 (MAP3K7IP2)
To be Published
|
|
2W2M
 
 | | WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
4GF5
 
 | | Crystal Structure of Calicheamicin Methyltransferase, CalS11 | | Descriptor: | CalS11, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Helmich, K.E, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: |
to be published
|
|
2N3T
 
 | |
