2IY8
 
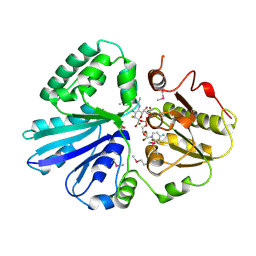 | | Crystal structure of the sialyltransferase PM0188 with CMP-3FNeuAc and lactose | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, PROTEIN PM0188, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
1EL9
 
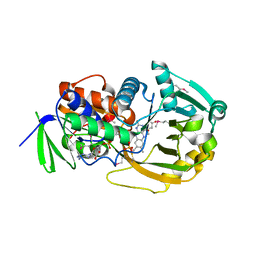 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLTHIO]ACETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
2W6W
 
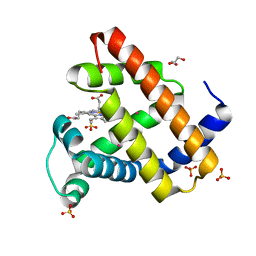 | | Crystal structure of recombinant Sperm Whale Myoglobin under 1atm of Xenon | | Descriptor: | GLYCEROL, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
6VZQ
 
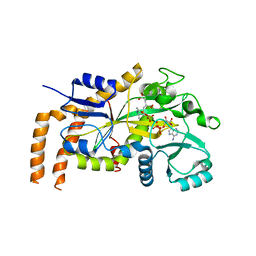 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZT
 
 | | TTLL6 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2GGI
 
 | |
1EL8
 
 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLSELENO]CETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
5IBF
 
 | |
5IBG
 
 | |
6VZV
 
 | | TTLL6 bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4AQ7
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and leucyl-adenylate analogue in the aminoacylation conformation | | Descriptor: | E. COLI TRNALEU UAA ISOACCEPTOR, LEUCINE, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Crepin, T, Vu, M.T, Lincecum Jr, T.L, Martinis, S.A, Cusack, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Dynamics of the Aminoacylation and Proofreading Functional Cycle of Bacterial Leucyl-tRNA Synthetase
Nat.Struct.Mol.Biol., 19, 2012
|
|
5CKK
 
 | |
3KBP
 
 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-10-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
5CIH
 
 | |
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6BCD
 
 | | Crystal structure of Rev7-K44A/R124A/A135D in complex with Rev3-RBM2 (residues 1988-2014) | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQU
 
 | | X-Ray crystal structure of the human carbonic anhydrase II adduct with a membrane-impermeant inhibitor | | Descriptor: | 4-[2-(2,4,6-triphenylpyridin-1-ium-1-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human carbonic anhydrase II adduct with 1-(4-sulfamoylphenyl-ethyl)-2,4,6-triphenylpyridinium perchlorate, a membrane-impermeant, isoform selective inhibitor.
J Enzyme Inhib Med Chem, 33, 2018
|
|
6S0M
 
 | |
1NZJ
 
 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
4OYX
 
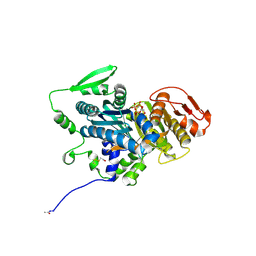 | | Human solAC Complexed with AMPCPP | | Descriptor: | Adenylate cyclase type 10, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
8XZV
 
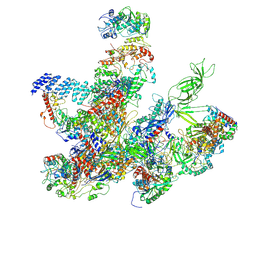 | | Architecture of the spinach plastid-encoded RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, G.-L, Yu, L.-J, Lu, C. | | Deposit date: | 2024-01-21 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Architecture of the spinach plastid-encoded RNA polymerase.
Nat Commun, 15, 2024
|
|
1AUX
 
 | |
5CIC
 
 | |
