8HUM
 
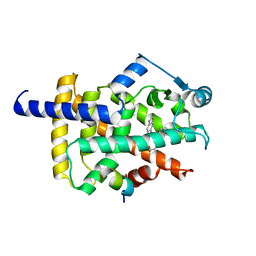 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUO
 
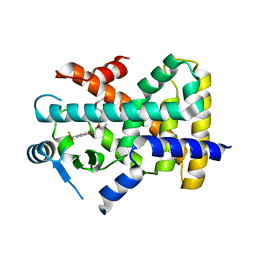 | | X-ray structure of human PPAR delta ligand binding domain-seladelpar co-crystals obtained by co-crystallization | | Descriptor: | Peroxisome proliferator-activated receptor delta, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUN
 
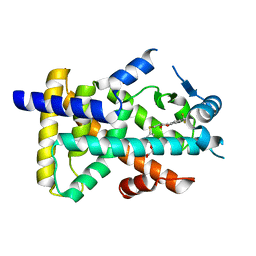 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5BRY
 
 | | HIV-1 wild Type protease with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
4J0R
 
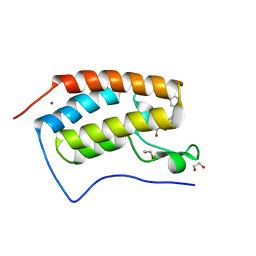 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(R)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
4J0S
 
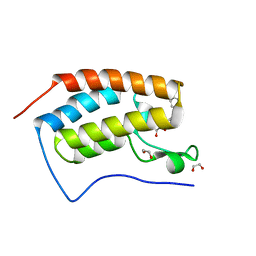 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
3CGU
 
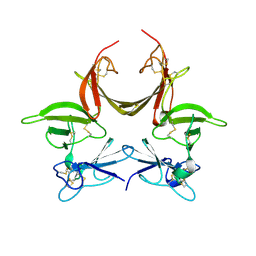 | |
7NP6
 
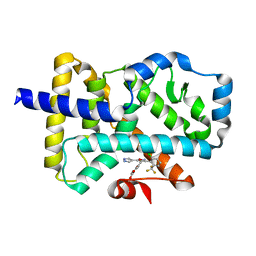 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM257 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrazol-4-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
9EX6
 
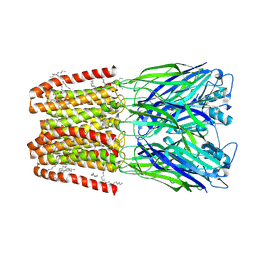 | | CryoEM structure of sTeLIC nanodisc in closed state | | Descriptor: | Cys-loop ligand-gated ion channel, DECANE, DODECANE, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-04-05 | | Release date: | 2025-03-26 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Vestibular modulation by stimulant derivatives in a pentameric ligand-gated ion channel.
Br.J.Pharmacol., 182, 2025
|
|
9EX4
 
 | | CryoEM structure of sTeLIC nanodisc in complex with fluorinated fos-choline-8 in open state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cys-loop ligand-gated ion channel, HEXANE, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-04-05 | | Release date: | 2025-03-26 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Vestibular modulation by stimulant derivatives in a pentameric ligand-gated ion channel.
Br.J.Pharmacol., 182, 2025
|
|
7NLD
 
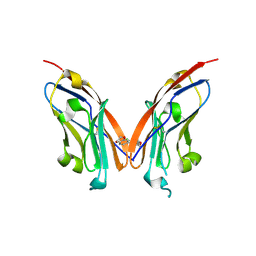 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | N-(2-((2'-chloro-3'-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-methoxy-[1,1'-biphenyl]-4-yl)(methyl)amino)ethyl)methanesulfonamide, Programmed cell death 1 ligand 1 | | Authors: | Sala, D, Magiera-Mularz, K, Muszak, D, Surmiak, E, Grudnik, P, Holak, T.A. | | Deposit date: | 2021-02-22 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
1QKT
 
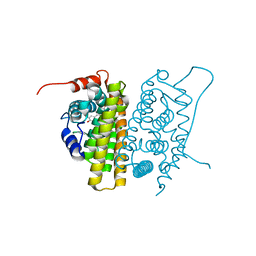 | | MUTANT ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Mutant Heralpha Ligand- Binding Domain Reveals Key Structural Features for the Mechanism of Partial Agonism
J.Biol.Chem., 276, 2001
|
|
5JON
 
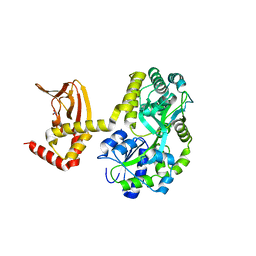 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
5UPQ
 
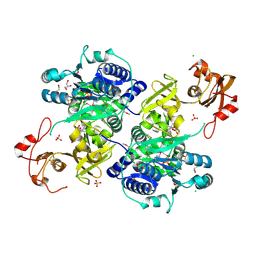 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP465 ligand | | Descriptor: | 5'-O-[(R)-{[(7beta,8alpha,9beta,10alpha,13alpha,16beta)-7,16-dihydroxy-18-oxokauran-18-yl]oxy}(hydroxy)phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
8GCB
 
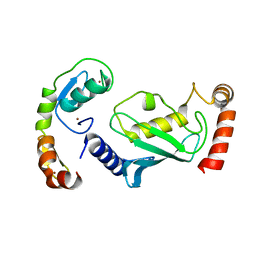 | | Structure of RNF125 in complex with a UbcH5b~Ub conjugate | | Descriptor: | E3 ubiquitin-protein ligase RNF125, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
8GBQ
 
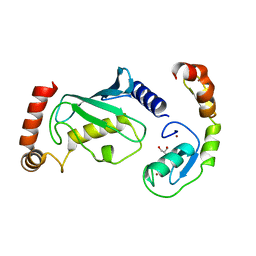 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
6U7O
 
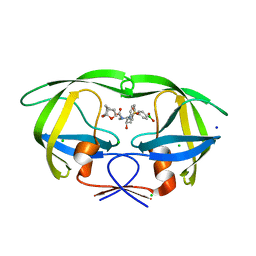 | | HIV-1 wild type protease with GRL-00819A, with phenyl-boronic-acid as P2'-ligand and with a 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
6U7P
 
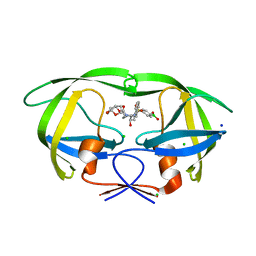 | | HIV-1 wild type protease with GRL-03119A, with phenyl-boronic-acid as P2'-ligand and with a hexahydro-4H-furo-pyran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
7W0W
 
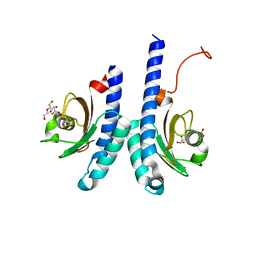 | | The novel membrane-proximal sensing mechanism in a broad-ligand binding chemoreceptor McpA of Bacillus velezensis | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Feng, H.C, Shen, Q.R, Zhang, R.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Signal binding at both modules of its dCache domain enables the McpA chemoreceptor of Bacillus velezensis to sense different ligands.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4F27
 
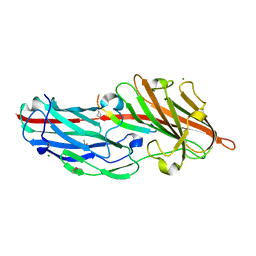 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Fibrinogen alpha chain | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
5ILC
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound 2, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5II3
 
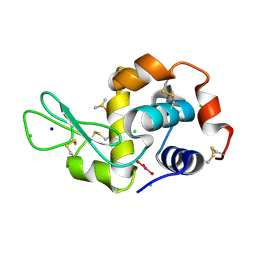 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 3, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5IHG
 
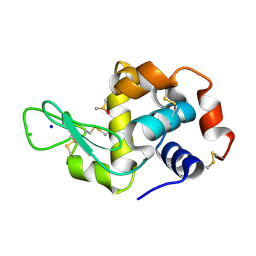 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound I, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-02-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
8TM0
 
 | | Preclinical Characterization of Pan-NKG2D Ligand-Binding NKG2D Receptor Decoys | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class I polypeptide-related sequence A, ... | | Authors: | Rupert, P.B, Strong, R. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Preclinical characterization of Pan-NKG2D ligand-binding NKG2D receptor decoys.
Heliyon, 10, 2024
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
